Abstract
Genes subjected to genomic imprinting are often associated with prenatal and postnatal growth. Furthermore, it has been observed that maternally silenced/paternally expressed genes tend to favour offspring growth, whilst paternally silenced/maternally expressed genes will restrict growth. One imprinted cluster in which this has been shown to hold true is the Gnas cluster; of the three proteins expressed from this cluster, two, Gsα and XLαs, have been found to affect postnatal growth in a number of different mouse models. The remaining protein in this cluster, NESP55, has not yet been shown to be involved in growth. We previously described a new mutation, Ex1A-T, which upon paternal transmission resulted in postnatal growth retardation due to loss of imprinting of Gsα and loss of expression of the paternally expressed XLαs. Here we describe maternal inheritance of Ex1A-T which gives rise to a small but highly significant overgrowth phenotype which we attribute to reduction of maternally expressed NESP55.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
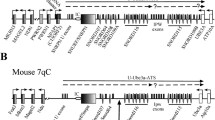
Genomic imprinting occurs in a subset of genes whereby gene expression is determined by the parent of origin. The imprinted Gnas cluster, located on chromosome 2 in mice and chromosome 20 in humans, contains transcripts that are maternally, paternally, and/or biallelically expressed (Peters et al. 1999) (Fig. 1). The Gnas cluster contains three protein-coding transcripts, Nesp, Gnasxl, and Gnas, which arise from three different promoters to give the proteins NESP55, XLαs, and Gsα (Peters and Williamson 2007). All three transcripts contain unique first exon(s) and all splice onto exon 2 of Gnas, from which point all the transcripts are identical. XLαs and Gsα, but not NESP55, can act as the α subunit of the heterotrimeric Gs protein, and both XLαs and Gsα have identical C-termini because their transcripts code protein through to exon 12 of Gnas. However, the NESP55 protein is generated exclusively from unique Nesp exon 2; the rest of the Nesp transcript, which is identical in sequence to exons 2–12 of Gnasxl and Gnas, comprises the 3′UTR.
Schematic diagram of the mouse Gnas cluster. Maternal (Mat) and paternal (Pat) transcripts are shown above and below the line, respectively. Maternal and paternal differentially methylated regions are shown above and below the line, respectively, and are designated with a “+.” Arrows show initiation and direction of transcription. Coding sections of the transcripts are shown in black whilst noncoding sections are shown in grey. Asterisk denotes that Gnas exon 2 can also be called Nesp exon 3. The transcript corresponding to the paternal Gnas allele is shown as a dotted line to denote that Gnas is paternally repressed in only some tissues. First exons of protein-coding transcripts are shown as filled boxes and first exons of noncoding transcripts are shaded boxes. Figure not to scale [adapted from Plagge et al. (2004) and Williamson et al. (2004)]
NESP55, or Neuroendocrine Secretory Protein of molecular weight 55,000, belongs to the granin family (Ischia et al. 1997). The granin family members are involved in endocrine and neuronal secretory pathways, and although the exact mechanisms of these proteins remain poorly understood, peptides derived from the granins have been shown to be involved in neuroendocrine, cardiovascular, endocrine, and inflammation systems. Many granins, including NESP55, have been shown to act as biomarkers for endocrine and neuroendocrine tumours (Bartolomucci et al. 2011). NESP55 is expressed in neural tissues such as the pons, midbrain, and hypothalamus; more specifically, high levels of expression are observed in the noradrenergic locus coeruleus, the dorsal raphe nucleus, the Edinger-Westphal nucleus, and the arcuate nucleus (Bauer et al. 1999; Plagge et al. 2005).
The Gnas cluster also contains two long noncoding RNA transcripts, Nespas, which transcribes antisense to Nesp, and Exon1A, whose first exon lies between the first exon of Gnasxl and Gnas and which also contains one unique first exon that splices onto exon 2 of Gnas (Liu et al. 2000; Wroe et al. 2000). Of these five transcripts, Nesp is exclusively maternally expressed; Nespas, Gnasxl, and Exon1A are exclusively paternally expressed; and Gnas is maternally expressed in a subset of tissues such as neonatal brown adipose tissue (BAT), the proximal renal tubules, and the paraventricular nucleus, but is biallelic in most other tissues (Chen et al. 2009; Kelsey et al. 1999; Liu et al. 2000; Peters et al. 1999; Williamson et al. 2004; Wroe et al. 2000; Yu et al. 1998).
The phenotypic effects of imprinted transcripts in the Gnas cluster broadly conform to the “kinship” or “parental conflict” theory of genomic imprinting. This states that there is a conflict between maternal and paternal genes in offspring in the demand for maternal resources, and it predicts that paternal alleles are more demanding while maternal alleles are less demanding (Moore and Haig 1991). Many imprinted genes have been shown to have major effects on postnatal growth and metabolism (Smith et al. 2006), and, overall, paternally expressed genes tend to favour growth while maternally expressed genes tend to restrict growth. In the case of the Gnas cluster, paternally expressed Gnasxl and maternally expressed Gnas act antagonistically and have opposite effects on body weight, adiposity, and metabolism. Thus, loss, or loss of function, of paternally expressed Gnasxl or increased expression of maternally expressed Gnas gives rise to mice that are growth restricted, lean, and hypermetabolic (Eaton et al. 2012; Kelly et al. 2009; Plagge et al. 2004; Xie et al. 2006; Yu et al. 1998, 2000), whereas loss of maternally expressed Gnas gives rise to mice that have increased body weights and are obese and hypometabolic (Chen et al. 2005; Germain-Lee et al. 2005; Kelly et al. 2009; Yu et al. 2000). Using the parental conflict theory as a guide, it might be expected that loss of maternally expressed Nesp may also affect size and adiposity and give rise to an overgrowth phenotype. However, in the Nesp knockout mouse, the only documented phenotype was abnormal behaviour to novel environments and a growth phenotype was not reported (Plagge et al. 2005).
Ex1A-T is a targeted mutation in which a polyadenylation (polyA) cassette was inserted into the Exon1A exon in order to truncate the Exon1A transcript upon paternal transmission (Eaton et al. 2012). We describe here that maternal inheritance of the Ex1A-T mutation results in increased body weight from prenatal stages onwards, truncation of the Nesp transcript after exon 2, and diminution of NESP55 protein, indicating that Nesp affects growth in accordance with the conflict hypothesis.
Methods
All mouse studies were carried out in accordance with the guidance issued by the Medical Research Council in “Responsibility in the use of animals in bioscience research” (May 2008) and under Home Office Project License No. 30/2526.
Mouse husbandry
The Ex1A-T transgenic line was created by insertion of a polyA cassette into the Exon1A exon as described previously (Eaton et al. 2012). Ex1A-T was maintained as a heterozygote on 129/SVEM. Toe clippings were taken at birth from pups; these served as both biopsies for genotyping and for identification. Mice were housed under specific pathogen-free conditions in individually ventilated cages, at 21 ± 2 °C, humidity of 55 ± 10 %, and subjected to a 12-h light/12-h dark cycle, in accordance with UK Home Office Welfare Guidelines. Mice had free access to water (25 ppm chloride) and food containing 11.5 kcal % fat, 23.93 kcal % protein, and 61.57 kcal % carbohydrate (SDS, RM3 diet).
Genotyping
Genomic DNA was extracted from mouse biopsies. Genotyping was performed by duplex PCR using primers described previously (Eaton et al. 2012).
RNA analysis
Total RNA was isolated using either the RNeasy or RNeasy lipid kits (Qiagen). Contaminating genomic DNA was removed on column with RNase-free DNase (Qiagen). cDNA was synthesised from 1 to 2 μg of RNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Quantitative (q)PCR was performed on an ABI Prism 7500 Fast system (Applied Biosystems) using either Fast SYBR Green PCR Master Mix (Applied Biosystems) or TaqMan® Fast Universal PCR Master Mix (Applied Biosystems), depending on the assay. Each reaction was done in triplicate with approximately 10 ng of cDNA per reaction. Samples were normalised to Gapdh levels. Analysis of data was carried out with ABI Prism 7500 system software (version 2.0.1). TaqMan probes are Gnas exon 1–2 (Mm00507037_m1, Life Technologies), as well as Nesp exon 1–2 and Gapdh, which were described previously (Williamson et al. 2011). Nesp SYBR primers are exon 2 fwd: AGATTCTCCTTGTTTTCATGGAT; exon 3 rev: GTTAAACCCATTAACATGCAGGA. Nespas SYBR primers are exon 1 fwd: AGATTTCATTTCCCAGAGATGCT; exon 2 rev: GGTTAGGCAGATCCGACTTGT. Gnasxl, Exon1A, and Gapdh SYBR primers were described previously (de Bovis et al. 2005; Eaton et al. 2012).
Western blotting
Total cell lysates were extracted using RIPA buffer (PBS with 1 % Nonidet P40, 0.5 % sodium deoxycholate, 0.1 % SDS). Cell lysate (50 μg) was loaded in each lane of a 4–12 % Bis–Tris NuPAGE gel (Invitrogen). Proteins were transferred to PVDF membrane (GE Healthcare), probed with specific antibodies followed by HPR-conjugated goat anti-rabbit secondary antibody (Sigma), and then detected using the ECL Plus Western Blotting Detection kit (GE Healthcare). The anti-rabbit NESP55 antibody (1:200) (Ischia et al. 1997) was directed against the C-terminal epitope GAIPIRRH. Anti-rabbit Gapdh (Sigma G9295) was used as a loading control. Films were then scanned by an Epson V750 Pro Scanner into.tif format and then quantitated using ImageJ software (http://rsb.info.nih.gov/ij/), following the methods outlined in the document http://www.lukemiller.org/ImageJ_gel_analysis.pdf.
Mouse weights
Individual mice of both sexes on a standard diet were weighed at birth and then weekly for 12 weeks. Each litter contained both wild-type and mutant mice. For each litter at each time point, the average wild-type weight of each sex was calculated and then each individual mouse’s weight within the litter was taken as a percentage of the average wild-type weight of corresponding sex.
DEXA analysis
At 12 weeks of age, male mice were weighed and given nonrecoverable general anaesthetic before scanning with a Lunar PIXImus Mouse Densitometer (Wipro, GE Healthcare). Fat mass, lean mass, bone mineral density, and body length were measured.
Metabolic caging
At 13 weeks of age, male mice were weighed and then individually housed in metabolic cages for 24 h, during which time they had free access to preweighed food and water. After the 24 h, the amount of food and water consumed was measured. After metabolic housing the mice were returned to their home cage.
Metabolic rate measurements
At 12 weeks of age, male mice were weighed and then housed individually in indirect calorimetry cages (Oxymax, Columbus Instruments) for 22 h. Mice had free access to food and water. Oxygen consumption, carbon dioxide consumption, respiratory exchange ratio, and heat production were analysed.
Plasma biochemistry
Blood samples were collected by terminal retro-orbital bleeds into lithium heparin Microvette tubes (Sarstedt UK) from 12-week-old male mice. Blood samples were centrifuged for 10 min at 5,000×g at 4 °C. Concentrations of total cholesterol, HDL cholesterol, LDL cholesterol, glucose, and triglycerides were determined on an AU680 clinical chemistry analyser (Beckman Coulter) using the manufacturer’s reagents and instructions. Fructosamine was also measured on the AU680 analyser, but with reagents and instructions respectively supplied by Randox, Alpha Laboratories, and Roche. Levels of plasma insulin, glucagon, and leptin were measured on a Bioplex 200 system (BioRad) using a multiplex endocrine assay kit (Merck Millipore).
Suckling activity
On postnatal day 5, suckling activity was assessed as described previously (Kelly et al. 2009).
Statistical methods
All comparisons were between cohorts of mutant and wild-type age-matched siblings. Comparisons between a given week’s weight and between signal quantification of NESP55 Western blots were made using an unpaired two-tailed Student’s t test, and comparisons between growth curves between genotypes were made using a repeated-measures ANOVA test. All other statistical comparisons were made using the nonparametric Mann–Whitney test. Ex1A-T/+ values are always given before +/+ values in the text.
Results
Ex1A-T/+ mice are larger than their wild-type siblings
Ex1A-T/+ (maternal allele listed first for all genotypes) mice were born at expected Mendelian ratios (51 % of 626 neonates). Ninety-seven percent of Ex1A-T/+ pups survived to weaning compared to 99 % of wild-type pups, which is not significantly different as measured by a χ2 test. As imprinted genes often affect growth, Ex1A-T/+ mice and their wild-type siblings were weighed weekly from birth. Only data from mice that survived to 12 weeks were considered, and weights are given as a percentage of their same-sex wild-type siblings (Fig. 2a). Ex1A-T/+ mice were found to be significantly (P < 0.001) heavier than their wild-type siblings by 4–8 %. Ex1A-T/+ mice were significantly (P = 0.0036) heavier at birth (104 %) than their wild-type siblings, indicating that the Ex1A-T/+ mutation regulates growth prenatally. The increase in weight peaked at 14 days of age at 108 % (P = 6 × 10−12), reached a minimum at 4 weeks of age of 104 % (P = 0.0014), and then increased to 106 % from 7 to 12 weeks of age (P = 4 × 10−7 to 1 × 10−8) when the study ended. Thus, the Ex1A-T/+ mutation gives rise to an overgrowth phenotype; mice show prenatal growth enhancement followed by an increased growth rate until postnatal day 14 (P14), then a growth retardation until weaning, and then a gradual increase in growth until the end of the study at 12 weeks.
The Ex1A-T/+ phenotype. a Growth curve of Ex1A-T/+ and their wild-type siblings of both sexes over 12 weeks. Wild-type sibling weights have been normalised to 1 at each time point and weights of the transgenic mice have been taken as a percentage of wild-type weights at each time point. Error bars indicate SEM. For any given time point, n = 90–112. For repeated-measures ANOVA test (0–12 weeks), n = 68, P < 0.001. b Growth curve of Ex1A-T/+, +/Ex1A-T, and Ex1A-T/Ex1A-T mice of both sexes over 12 weeks. Wild-type sibling weights have been normalised to 1 at each time point and weights of the transgenic mice have been taken as a percentage of sibling wild-type weights at each time point. Error bars indicate SEM. For any given time point, n = 14–20 +/Ex1A-T, n = 97–109 Ex1A-T/+, n = 20–23 Ex1A-T/Ex1A-T. For repeated-measure ANOVA test of +/Ex1A-T versus Ex1A-T/Ex1A-T (3–12 weeks), n = 12–19, P < 0.05. For repeated-measure ANOVA test of Ex1A-T/Ex1A-T versus Ex1A-T/+ (0–12 weeks), n = 16–68, P < 0.001. +/Ex1A-T curve taken from Eaton et al. (2012). c Organ weights of 12-week-old male mice. Error bars indicate SEM, n = 6–7, **P < 0.05, ***P < 0.01. d Total fat and lean mass of 12-week-old male mice as analysed by DEXA. Error bars indicate SEM, n = 6–7, ***P < 0.01. e Bone mineral density of 12-week-old male mice as analysed by DEXA. Error bars indicate SEM, n = 6–7, P = 0.010
We also intercrossed Ex1A-T heterozygotes to generate Ex1A-T/Ex1A-T homozygotes. As described previously (Eaton et al. 2012), +/Ex1A-T mice are smaller than wild-type siblings due to underexpression of the paternal Gnasxl and overexpression of the maternal Gnas transcripts. When the maternal Ex1A-T allele is carried with the paternal Ex1A-T allele, the resulting animals have an intermediate weight phenotype (Fig. 2b), demonstrating the antagonistic relationship between paternally expressed and maternally expressed genes.
Next we sought to determine what was causing the Ex1A-T/+ mice to be heavier. Organ weights from 12-week-old male mice were measured (Fig. 2c). The weights of brain, liver, and kidneys (but not BAT, white adipose tissue (WAT), heart, lung, or spleen) of Ex1A-T/+ were found to be slightly, but significantly, heavier than their wild-type siblings. However, when the organ weight was taken as a percentage of body weight, only the kidneys were found to be significantly heavier (P = 0.022 and 0.008, left and right kidneys, respectively), while the lungs were found to be significantly lighter (P = 0.022). Lean and fat mass were also assessed in 12-week-old male mice by DEXA (Fig. 2d). A significant (P = 0.005) increase in total lean mass (2.55 g) and an increase in fat mass (0.55 g), which was not significant (P = 0.3) were observed in the Ex1A-T/+ mice. No difference was observed in either lean or fat mass when compared to body weight (fat mass: 19.0 vs. 19.0 %, P = 0.95; lean mass: 79.7 vs. 79.1 %, P = 0.53). This indicates that the increase in body weight of Ex1A-T/+ mice is predominantly due to an increase in lean, not fat, mass, and this increase is in proportion to the whole mouse. Bone mineral density (BMD) was also assessed by DEXA in 12-week-old male mice. Ex1A-T/+ mice were found to have a significantly greater BMD than wild-type (Fig. 2E). However, Ex1A-T/+ mice did not have an increase in either bone or body length (femur length: 15.22 vs. 15.24 mm, P = 0.85, n = 3–4; body length: 11.1 vs. 10.9 cm, P = 0.38, n = 6). As a consequence of increased body weight and unchanged body length, the Ex1A-T/+ mice were found to have a significantly increased BMI (0.249 vs. 0.226, P = 0.037, n = 6).
Metabolic rate was measured in 12-week-old male mice and no significant difference in oxygen consumption and carbon dioxide output was observed (data not shown). Food intake in Ex1A-T/+ mice was assessed in terms of body mass at 5 days and 12 weeks and no significant difference (although there was a trend to increase) in food intake was observed (5 days: 2.25 % vs. 2.01 %, P = 1, n = 18–20; 12 weeks: 13.7 % vs. 12.6 %, P = 1, n = 6–3). There was also no significant difference in total food intake at 12 weeks (3.47 vs. 3.02 g, P = 0.381, n = 6–3). Comparison of male Ex1A-T/+ and wild-type siblings at 12 weeks showed no significant differences in plasma glucose, fructosamine, insulin, glucagon, leptin, total cholesterol, HDL cholesterol, LDL cholesterol, or triglyceride level levels (results not shown).
Maternal transmission of Ex1A-T results in truncation of Nesp and reduction in NESP55 protein
The Ex1A-T targeted mutation contains a polyadenylation (polyA) cassette inserted into the end of the Exon1A (first) exon and was originally designed to truncate the Exon1A transcript upon paternal transmission (Eaton et al. 2012). It was also found to truncate the Gnasxl transcript upon paternal transmission, as the polyA cassette lies in the first intron of Gnasxl. On maternal transmission the only transcript that should contain the polyA cassette in either an intron or an exon is Nesp, with the polyA cassette in its second intron. Thus, we hypothesised that maternal inheritance of Ex1A-T should result in truncation of the Nesp transcript after exon 2. We therefore analysed Nesp expression levels both upstream of the truncation (Nesp exons 1–2) and across the truncation (Nesp exon 2–Nesp exon 3; note that Nesp exon 3 is also Gnas exon 2) by quantitative (q)PCR in brain, BAT, and in some cases the pituitary of newborn Ex1A-T/+ and wild-type littermates (Fig. 3a, b). These tissues were selected because many Gnas cluster transcripts are highly expressed in the brain and BAT of newborns; moreover, Gnas is imprinted in the BAT of newborns and pituitary but not whole brain. We found expression equivalent to that of wild-type in the Ex1A-T/+ mice between Nesp exons 1 and 2, but a drastic reduction in expression between Nesp exons 2 and 3. This is consistent with truncation of the Nesp transcript at the Exon1A exon, between exons 2 and 3 of Nesp.
Transcript analysis of Ex1A-T/+ mice. a TaqMan qPCR analysis of upstream Nesp transcript in newborn tissues. This probe is specific for Nesp exon 1–2, upstream of the polyA cassette. Expression was normalised to Gapdh. Error bars indicate SEM, n = 7–9, P = 0.68 for BAT, 0.11 for brain. b SYBR qPCR analysis of truncated Nesp transcript in newborn tissues. Primers were designed at the end of Nesp exon 2 and at the start of Nesp exon 3 (Gnas exon 2) to analyse transcript downstream of the truncation. Expression was normalised to Gapdh, error bars indicate SEM, n = 6–9, P ≤ 0.0012. c TaqMan qPCR analysis of Gnas transcript in newborn tissues. Expression was normalised to Gapdh, error bars indicate SEM, n = 7–9 for BAT and brain, n = 5 for pituitary; P = 0.47 for BAT, 0.61 for brain, 0.076 for pituitary. d SYBR qPCR analysis of Exon1A transcript in newborn tissues. Primers were designed at the end of the Exon1A exon and at the start of Exon1A exon 2 (Gnas exon 2). Expression was normalised to Gapdh, error bars indicate SEM, n = 7–9, P = 0.61 for BAT, 0.68 for brain. e SYBR qPCR analysis of Gnasxl transcript in newborn tissues. Primers were designed at the end of the Gnasxl exon and at the start of Gnasxl exon 2 (Gnas exon 2). Expression was normalised to Gapdh, error bars indicate SEM, n = 7–9, P = 0.76 for BAT, 0.76 for brain. f SYBR qPCR analysis of Nespas transcript in newborn tissues. Primers were designed at the end of the Nespas exon 1 and at the start of Nespas exon 2. Expression was normalised to Gapdh, error bars indicate SEM, n = 2–4, P = 0.8 for BAT, 0.27 for brain. g Western blot of NESP55 and Gapdh loading control in newborn pituitary; 13–14 pituitaries were pooled for each genotype and the blot is representative of at least two replicates. h Signal quantification of NESP55 Western blots, represented in (g). Expression was normalised to Gapdh, error bars indicate SEM, n = 2, P = 0.0083 (assuming a normal distribution)
Transcription of Nesp is required for the establishment of methylation marks on the maternal allele and imprinted expression of transcripts in the Gnas cluster (Chotalia et al. 2009). A previous truncation of the Nesp transcript within its second exon resulted in disruption of methylation at the Gnasxl differentially methylated region (DMR) and the Exon1A DMR, disrupting expression of downstream Gnas cluster transcripts (Chotalia et al. 2009). In Ex1A-T/+, Nesp should be transcribed through both DMRs as the polyA cassette is inserted close to the 3′ end of the second DMR at Exon1A and so the methylation of the DMRs and expression of the other Gnas cluster transcripts should be unaffected. We analysed all major Gnas cluster transcripts, i.e., Gnas, Exon1A, Gnasxl, and Nespas, by qPCR (Fig. 3c–f). None of these transcripts showed any change in expression levels between Ex1A-T/+ and wild-type littermates, confirming that Nesp is the only Gnas cluster transcript affected by this mutation.
Only exon 2 of Nesp codes for protein, and the transcript downstream of exon 2 is 3′UTR. Therefore, we next sought to determine if NESP55 protein was affected in Ex1A-T/+. NESP55 levels in neonatal pituitary were analysed by Western blotting using an antibody to NESP55 directed against the C-terminal epitope GAIPIRRH (Ischia et al. 1997). Neonatal pituitary was chosen because Nesp expression was highest in this tissue. Interestingly, NESP55 protein was found to be reduced upon truncation of the Nesp transcript (Fig. 3g). Quantification of the Western blot showed a fivefold reduction in NESP55 protein in the Ex1A-T/+ mice (Fig. 3h). We hypothesise that truncation of the 3′UTR of Nesp affects translation of the Nesp transcript, resulting in a reduction of NESP55 at the protein level.
Discussion
Nesp is the candidate for the Ex1A-T/+ phenotype
On maternal inheritance of the Gnas cluster mutation Ex1A-T/+, mice display a relatively mild but highly significant overgrowth phenotype with a 4–8 % increase in body weight compared to their wild-type siblings. This increase in body weight can be at least partly attributed to an increase in the absolute weight of lean tissues. These mice also show an increase in bone mineral density at 12 weeks of age, but not an increase in bone length or body length. The increase in weight does not appear to be due to an increase in fat mass, food intake, or a reduction in metabolic rate.
In Ex1A-T/+, insertion of a polyA cassette on the maternal allele results in truncation of the 3′UTR of the maternal Nesp transcript. The only transcripts from the Gnas cluster that are maternally expressed are Nesp and Gnas itself, and as the start site of the Gnas transcript is downstream of the polyA cassette, the Gnas transcript was neither expected nor found to be truncated. There is also no evidence in the present study or from Eaton et al. (2012) that insertion of the polyA cassette itself reduces the level of Gnas expression. Furthermore, Ex1A-T/+ mice do not show the increased adiposity and decreased metabolic rate that is characteristic of mutants with reduced Gnas (Chen et al. 2005; Kelly et al. 2009; Xie et al. 2008). Analysis of other Gnas cluster transcripts confirmed that Nesp was the only transcript in the Gnas cluster that was altered in the Ex1A-T/+ mice and NESP55 protein appeared to be much reduced. Thus, Nesp is the logical candidate for the phenotype in Ex1A-T/+.
The 3′UTR is known to contain sequence elements that regulate the stability, translational control, and subcellular localisation of an mRNA. These sequence elements include miRNA binding sites, AU-rich elements, and polyA sites, and the latter have only recently been thought to significantly contribute to gene regulation. Investigations into the outcomes of alternative polyA site usage in the 3′UTR have revealed that different lengths of the 3′UTR can alter the amount of protein made (Di Giammartino et al. 2011). This can occur through loss of miRNA or other regulatory sequences. In general, longer 3′UTRs will contain more of these regulatory sequences and thus be prone to negative regulation. However, alternative polyA usage can also affect the amount of protein generated by an mRNA. Yu et al. (2006) identified a novel shorter Bzw1 transcript in mice that shared the same coding region as the longer transcript but had a shorter 3′UTR due to an alternative polyA site (Yu et al. 2006). Labelled constructs containing each of these transcripts were transfected into HEK-293 cells and were found to produce equivalent amounts of mRNA. Interestingly, the transcript with the shorter 3′UTR was found to produce significantly less protein, indicating reduced translation efficiency. In a related study, Pinto et al. (2011) showed that the two alternative 3′UTR polyA site transcripts of the Drosophila gene polo produced equivalent amounts of mRNA and had equally stable mRNA, but that the shorter transcript produced significantly (threefold) less protein (Pinto et al. 2011). Thus, we hypothesise that, similar to the previous two studies, shortening of the 3′UTR of Nesp results in reduced translational efficiency and reduced protein expression and that reduction in NESP55 leads to the Ex1A-T/+ phenotype.
A truncation of the Nesp transcript has been previously described. Chotalia et al. (2009) showed that transcription of Nesp across the germline DMRs at Gnasxl/Nespas and Exon1A (Fig. 1) was required for the establishment of methylation. A polyA cassette identical to the one in the present study was used to truncate the Nesp transcript upstream of both the Gnasxl/Nespas DMR and the Exon1A DMR. As a consequence, methylation was lost on the maternal allele at both DMRs, resulting in ectopic expression of Gnasxl, Nespas, and Exon1A transcripts from the normally silent maternal allele, downregulation of the imprinted Gnas transcript, and very poor postnatal survival. In contrast, the Ex1A-T/+ truncation of Nesp occurs downstream of both the Gnasxl/Nespas and Exon1A DMRs, and no change in expression was observed in any of the downstream Gnas cluster transcripts Gnasxl, Nespas, Exon1A, and Gnas. These findings imply that methylation of the Gnasxl/Nespas DMR and the Exon1A DMR is unaffected in Ex1A-T/+ so that transcription of Nesp through the DMRs rather than the full length transcription is sufficient for methylation. These results are consistent with the hypothesis proposed by Chotalia et al. (2009).
Reciprocal phenotypic effects
We previously described both paternal transmission of the Ex1A-T mutation and paternal transmission of the Ex1A-T-CON mutation (Eaton et al. 2012). The Ex1A-T mutation resulted in reduced expression of the paternally expressed Gnas cluster transcript Gnasxl and an increase in the maternally expressed Gnas transcript, while the Ex1A-T-CON mutation resulted in increased maternally expressed Gnas transcript only. Both these mutations presented with growth retardation which was present from birth. Interestingly, the phenotype observed upon maternal transmission of the Ex1A-T allele is in many ways the reciprocal in regard to growth to that of paternal inheritance of Ex1A-T or Ex1A-T-CON, despite different unrelated proteins being affected. In addition, the shape of the growth curve of the Ex1A-T/+ mice is the mirror image of that of the +/Ex1A-T and +/Ex1A-T-CON mice. All three mutations present with a difference in birth weight. The largest difference in weight between the mutants and their wild-type siblings is at 2 weeks of age. The mice then recover somewhat and then the weight stabilises (compared to that of wild-type) at about 7 weeks of age. Both Gsα and XLαs have been shown to regulate preweaning growth and metabolism in a number of different mouse models (Cattanach et al. 2000; Eaton et al. 2012; Plagge et al. 2004; Yu et al. 2000).
The Ex1A-T/+ mice were observed to have an increase in bone mineral density; once again this is the reciprocal of what was observed in the +/Ex1A-T mice (but not in the case of the +/Ex1A-T-CON). However, we proposed that the reduction in BMD in the +/Ex1A-T mice was linked to their reduced fat mass and was regulated by the sympathetic nervous system (SNS), which has been shown to regulate both bone and adipose formation (Eaton et al. 2012). Ex1A-T/+ mice do not have a corresponding increase in fat mass, and as NESP55 is not associated with G proteins it is unlikely to be able to directly modulate SNS activity. Thus, we hypothesise that the mechanisms of regulation of bone density differ for NESP55 and XLαs.
The Gnas cluster and growth
All three protein-coding genes from the Gnas locus, i.e., Gnas, Gnasxl, and Nesp, have now been implicated in preweaning growth. Moreover, mice that carry both the paternal Ex1A-T and maternal Ex1A-T alleles have an intermediate growth phenotype between Ex1A-T/+ and +/Ex1A-T, illustrating the antagonistic relationship between underexpression of paternal XLαs combined with overexpression of maternal Gsα and underexpression of maternal NESP55. Rescue of an imprinted phenotype by an oppositely imprinted gene has also been seen with crosses of Igf2r/+ to +/Igf2 which result in rescue of the perinatal lethality and overgrowth phenotype (Ludwig et al. 1996). The antagonistic relationship of paternally expressed XLαs and maternally expressed Gsα is well studied. XLαs and Gsα have been shown to have opposing effects on growth, adiposity, and metabolic rate (Plagge et al. 2004; Xie et al. 2006; Yu et al. 2000), as well as opposite molecular effects by which XLαs represses but Gsα activates SNS activity (Plagge et al. 2004; Xie et al. 2006; Yu et al. 2000). Whereas cAMP signalling is stimulated by Gsα, there are reports that it is inhibited by XLαs (Plagge et al. 2004; Weinstein et al. 2004; Xie et al. 2006). On the other hand, the mechanisms of action of NESP55 (and other members of the granin family) are poorly understood. However, granins have been shown to be involved in endocrine and neuronal secretory pathways (Bartolomucci et al. 2011), and NESP55 is considered to be a marker for the constitutive secretory pathway (Eder et al. 2004; Fischer-Colbrie et al. 2002). Given that XLαs/Gsα and NESP55 are involved in different pathways, one would predict the ways they mediate pre- and postnatal growth to differ as well. Thus, the nature of the antagonistic relationship between NESP55 and XLαs remains obscure.
This reciprocal pattern of growth for maternal and paternal transcripts is consistent with the parental conflict theory of the evolution of genomic imprinting. Loss of maternally expressed Nesp results in growth promotion, whereas loss of paternally expressed Gnasxl results in restricted growth.
Although many maternally expressed proteins have been shown to affect embryonic or placental growth, only a handful with a postnatal overgrowth phenotype upon loss or reduction of the maternal transcript have been described, including Gsα, Grb10, and Atp10c (Charalambous et al. 2007). Similar to Ex1A-T/+, mice with loss of maternal Grb10 show both a prenatal and a postnatal overgrowth phenotype. However, unlike the Ex1A-T/+ mice, this overgrowth does not appear to peak at 2 weeks of age and the Grb10/+ mice were found to have reduced WAT (Smith et al. 2007). Pre- and postnatal overgrowth is also a characteristic of the human disorder Beckwith–Wiedemann syndrome, which is caused by loss of maternally expressed genes at 11p15 (Ishida and Moore 2013).
Until now NESP55 has not been thought to be involved in growth and metabolic effects (Plagge et al. 2005). Here we have shown, using Ex1A-T/+ mice, that NESP55 may indeed play a role in growth.
References
Bartolomucci A, Possenti R, Mahata SK, Fischer-Colbrie R, Loh YP, Salton SR (2011) The extended granin family: structure, function, and biomedical implications. Endocr Rev 32:755–797
Bauer R, Ischia R, Marksteiner J, Kapeller I, Fischer-Colbrie R (1999) Localization of neuroendocrine secretory protein 55 messenger RNA in the rat brain. Neuroscience 91:685–694
Cattanach BM, Peters J, Ball S, Rasberry C (2000) Two imprinted gene mutations: three phenotypes. Hum Mol Genet 9:2263–2273
Charalambous M, da Rocha ST, Ferguson-Smith AC (2007) Genomic imprinting, growth control and the allocation of nutritional resources: consequences for postnatal life. Curr Opin Endocrinol Diabetes Obes 14:3–12
Chen M, Gavrilova O, Liu J, Xie T, Deng C, Nguyen AT, Nackers LM, Lorenzo J, Shen L, Weinstein LS (2005) Alternative Gnas gene products have opposite effects on glucose and lipid metabolism. Proc Natl Acad Sci USA 102:7386–7391
Chen M, Wang J, Dickerson KE, Kelleher J, Xie T, Gupta D, Lai EW, Pacak K, Gavrilova O, Weinstein LS (2009) Central nervous system imprinting of the G protein G(s)alpha and its role in metabolic regulation. Cell Metab 9:548–555
Chotalia M, Smallwood SA, Ruf N, Dawson C, Lucifero D, Frontera M, James K, Dean W, Kelsey G (2009) Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev 23:105–117
de Bovis B, Derouet D, Gauchat JF, Elson G, Gascan H, Delapeyriere O (2005) clc is co-expressed with clf or cntfr in developing mouse muscles. Cell Commun Signal 3:1
Di Giammartino DC, Nishida K, Manley JL (2011) Mechanisms and consequences of alternative polyadenylation. Mol Cell 43:853–866
Eaton SA, Williamson CM, Ball ST, Beechey CV, Moir L, Edwards J, Teboul L, Maconochie M, Peters J (2012) New mutations at the imprinted Gnas cluster show gene dosage effects of Gsalpha in postnatal growth and implicate XLalphas in bone and fat metabolism but not in suckling. Mol Cell Biol 32:1017–1029
Eder S, Leierer J, Klimaschewski L, Wilhelm A, Volknandt W, Laslop A, Fischer-Colbrie R (2004) Secretion and molecular forms of NESP55, a novel genomically imprinted neuroendocrine-specific protein from AtT-20 cells. Neurosignals 13:298–307
Fischer-Colbrie R, Eder S, Lovisetti-Scamihorn P, Becker A, Laslop A (2002) Neuroendocrine secretory protein 55: a novel marker for the constitutive secretory pathway. Ann N Y Acad Sci 971:317–322
Germain-Lee EL, Schwindinger W, Crane JL, Zewdu R, Zweifel LS, Wand G, Huso DL, Saji M, Ringel MD, Levine MA (2005) A mouse model of Albright hereditary osteodystrophy generated by targeted disruption of exon 1 of the Gnas gene. Endocrinology 146:4697–4709
Ischia R, Lovisetti-Scamihorn P, Hogue-Angeletti R, Wolkersdorfer M, Winkler H, Fischer-Colbrie R (1997) Molecular cloning and characterization of NESP55, a novel chromogranin-like precursor of a peptide with 5-HT1B receptor antagonist activity. J Biol Chem 272:11657–11662
Ishida M, Moore GE (2013) The role of imprinted genes in humans. Mol Aspects Med 34(4):826–840
Kelly ML, Moir L, Jones L, Whitehill E, Anstee QM, Goldin RD, Hough A, Cheeseman M, Jansson JO, Peters J, Cox RD (2009) A missense mutation in the non-neural G-protein alpha-subunit isoforms modulates susceptibility to obesity. Int J Obes (Lond) 33:507–518
Kelsey G, Bodle D, Miller HJ, Beechey CV, Coombes C, Peters J, Williamson CM (1999) Identification of imprinted loci by methylation-sensitive representational difference analysis: application to mouse distal chromosome 2. Genomics 62:129–138
Liu J, Yu S, Litman D, Chen W, Weinstein LS (2000) Identification of a methylation imprint mark within the mouse Gnas locus. Mol Cell Biol 20:5808–5817
Ludwig T, Eggenschwiler J, Fisher P, D’Ercole AJ, Davenport ML, Efstratiadis A (1996) Mouse mutants lacking the type 2 IGF receptor (IGF2R) are rescued from perinatal lethality in Igf2 and Igf1r null backgrounds. Dev Biol 177:517–535
Moore T, Haig D (1991) Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet 7:45–49
Peters J, Williamson CM (2007) Control of imprinting at the Gnas cluster. Epigenetics 2:207–213
Peters J, Wroe SF, Wells CA, Miller HJ, Bodle D, Beechey CV, Williamson CM, Kelsey G (1999) A cluster of oppositely imprinted transcripts at the Gnas locus in the distal imprinting region of mouse chromosome 2. Proc Natl Acad Sci USA 96:3830–3835
Pinto PA, Henriques T, Freitas MO, Martins T, Domingues RG, Wyrzykowska PS, Coelho PA, Carmo AM, Sunkel CE, Proudfoot NJ, Moreira A (2011) RNA polymerase II kinetics in polo polyadenylation signal selection. EMBO J 30:2431–2444
Plagge A, Gordon E, Dean W, Boiani R, Cinti S, Peters J, Kelsey G (2004) The imprinted signaling protein XL alpha s is required for postnatal adaptation to feeding. Nat Genet 36:818–826
Plagge A, Isles AR, Gordon E, Humby T, Dean W, Gritsch S, Fischer-Colbrie R, Wilkinson LS, Kelsey G (2005) Imprinted Nesp55 influences behavioral reactivity to novel environments. Mol Cell Biol 25:3019–3026
Smith FM, Garfield AS, Ward A (2006) Regulation of growth and metabolism by imprinted genes. Cytogenet Genome Res 113:279–291
Smith FM, Holt LJ, Garfield AS, Charalambous M, Koumanov F, Perry M, Bazzani R, Sheardown SA, Hegarty BD, Lyons RJ, Cooney GJ, Daly RJ, Ward A (2007) Mice with a disruption of the imprinted Grb10 gene exhibit altered body composition, glucose homeostasis, and insulin signaling during postnatal life. Mol Cell Biol 27:5871–5886
Weinstein LS, Liu J, Sakamoto A, Xie T, Chen M (2004) Minireview: GNAS: normal and abnormal functions. Endocrinology 145:5459–5464
Williamson CM, Ball ST, Nottingham WT, Skinner JA, Plagge A, Turner MD, Powles N, Hough T, Papworth D, Fraser WD, Maconochie M, Peters J (2004) A cis-acting control region is required exclusively for the tissue-specific imprinting of Gnas. Nat Genet 36:894–899
Williamson CM, Ball ST, Dawson C, Mehta S, Beechey CV, Fray M, Teboul L, Dear TN, Kelsey G, Peters J (2011) Uncoupling antisense-mediated silencing and DNA methylation in the imprinted Gnas cluster. PLoS Genet 7:e1001347
Wroe SF, Kelsey G, Skinner JA, Bodle D, Ball ST, Beechey CV, Peters J, Williamson CM (2000) An imprinted transcript, antisense to Nesp, adds complexity to the cluster of imprinted genes at the mouse Gnas locus. Proc Natl Acad Sci USA 97:3342–3346
Xie T, Plagge A, Gavrilova O, Pack S, Jou W, Lai EW, Frontera M, Kelsey G, Weinstein LS (2006) The alternative stimulatory G protein alpha-subunit XLalphas is a critical regulator of energy and glucose metabolism and sympathetic nerve activity in adult mice. J Biol Chem 281:18989–18999
Xie T, Chen M, Gavrilova O, Lai EW, Liu J, Weinstein LS (2008) Severe obesity and insulin resistance due to deletion of the maternal Gsalpha allele is reversed by paternal deletion of the Gsalpha imprint control region. Endocrinology 149:2443–2450
Yu S, Yu D, Lee E, Eckhaus M, Lee R, Corria Z, Accili D, Westphal H, Weinstein LS (1998) Variable and tissue-specific hormone resistance in heterotrimeric Gs protein alpha-subunit (Gsalpha) knockout mice is due to tissue-specific imprinting of the Gsalpha gene. Proc Natl Acad Sci USA 95:8715–8720
Yu S, Gavrilova O, Chen H, Lee R, Liu J, Pacak K, Parlow AF, Quon MJ, Reitman ML, Weinstein LS (2000) Paternal versus maternal transmission of a stimulatory G-protein alpha subunit knockout produces opposite effects on energy metabolism. J Clin Invest 105:615–623
Yu M, Sha H, Gao Y, Zeng H, Zhu M, Gao X (2006) Alternative 3′UTR polyadenylation of Bzw1 transcripts display differential translation efficiency and tissue-specific expression. Biochem Biophys Res Commun 345:479–485
Acknowledgments
We thank Denise Barlow and Frank Sleutels for providing a cloned fragment of the rabbit β-globin gene. We also thank Bruce Cattanach, Chris Williamson, and Simon Ball for helpful discussions; Joseph Maynard for image analysis; staff of the Mary Lyon Centre (MLC) for animal husbandry, in particular Lynn Jones, Diane Napper, Jackie Harrison, Kelly Hunt, Lucie Vizor, and Sara Wells; the MLC transgenic service; the MLC clinical pathology service; and the MLC necropsy service.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Eaton, S.A., Hough, T., Fischer-Colbrie, R. et al. Maternal inheritance of the Gnas cluster mutation Ex1A-T affects size, implicating NESP55 in growth. Mamm Genome 24, 276–285 (2013). https://doi.org/10.1007/s00335-013-9462-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-013-9462-2