Abstract
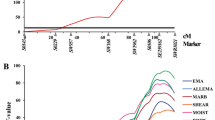
A crossed population between Iberian × Landrace pigs consisting of 321 F2, 87 F3, and 85 backcross individuals has been analyzed to refine the number and positions of quantitative trait loci (QTL) affecting shape, growth, fatness, and meat quality traits in SSC4. A multitrait multi-QTL approach has been used. Our results suggest that carcass length and shoulder weight are affected by two loci. The first one, close to the AFABP gene, has a very strong pleiotropic effect on fatness, whereas the second one, in the interval between S0073 and S0214, also affects live weight, although to a lesser extent. This latter QTL would correspond to the FAT1 locus described initially in pigs. It seems that SSC4’s loci play an important role in redistributing total weight, and the Landrace allele increases shoulder weight and carcass length much more than ham or total weight. Furthermore, there is also strong evidence of additional loci influencing pH and color in more distant, telomeric positions.


Similar content being viewed by others
References
Andersson L, Haley CS, Ellegren H, Knott SA, Johansson M, et al. (1994) Genetic mapping of quantitative trait loci for growth and fatness in pigs. Science 263, 1771–1774
Ausubel F, Brent R, Kingston R, Moore D, Seidman J, et al. (1987) Current protocols in molecular biology. (New York: Greene Publishing Associates and Wiley-Interscience)
Bidanel JP, Milan D, Iannuccelli N, Amigues Y, Boscher MY, et al. (2001) Detection of quantitative trait loci for growth and fatness in pigs. Genet Sel Evol 33, 289–309
Bidanel JP, Rothschild M (2002) Current status of quantitative trait locus mapping in pigs. Pig News and Information 23, 39–54
Clop A, Cercos A, Tomas A, Pérez–Enciso M, Varona L, et al. (2002) Assignment of the 2,4-dienoyl-CoA reductase (DECR) gene to porcine chromosome 4. Anim Genet 33, 164–165
Darvasi A (1998) Experimental strategies for the genetic dissection of complex traits in animal models. Nat Genet 18, 19–24
De Koning DJ, Janss LL, Rattink AP, van Oers PA, de Vries BJ, et al. (1999) Detection of quantitative trait loci for back fat thickness and intramuscular fat content in pigs (Sus scrofa). Genetics 152, 1679–1690
De Koning DJ, Rattink AP, Harlizius B, Groenen MAM, Brascamp EW, et al. (2001) Detection and characterization of quantitative trait loci for growth and reproduction traits in pigs. Livest Prod Sci 72, 185–198
Frary A, Fritz L, Tanksley S (2004) A comparative study of the genetic bases of natural variation in tomato leaf, sepal, and petal morphology. Theor Appl Genet 109, 523–533
Gilbert H, Le Roy P (2003) Comparison of three multitrait methods for QTL detection. Genet Sel Evol 35; 281–304
Green P, Falls K, Crooks S (1990) Documentation for CRIMAP. Unpublished mimeo. Available at: http ://biobase.dk/Embnetut/Crimap/
Iannuccelli N, Woloszyn N, Arhainx J, Gellin J, Milan D (1996) GEMMA: a database to manage and automate microsatellite genotyping. In Proceeding of the International Society of Animal Genetics Conference, Tours, France. (Oxford, UK: Blackwell) p 88
Jonsson P (1975) Methods of pig improvement through breeding in the European countries: A review. Livest Prod Sci 2, 1–28
Klingenberg CP, Leamy LJ, Routman EJ, Cheverud JM (2001) Genetic architecture of mandible shape in mice: Effects of quantitative trait loci analyzed by geometric morphometrics. Genetics 157, 785–802
Knott SA, Haley CS (2000) Multitrait least squares for quantitative trait loci detection. Genetics 156, 899–911
Knott SA, Marklund L, Haley CS, Andersson K, Davies W, et al. (1998) Multiple marker mapping of quantitative trait loci in a cross between outbred wild boar and large white pigs. Genetics 149, 1069–1080
Knott SA, Nystrom PE, Anderssoneklund L, Stern S, Marklund L, et al. (2002) Approaches to interval mapping of QTL in a multigeneration pedigree: the example of porcine chromosome 4. Anim Genet 33, 26– 32
Marklund L, Nystrom PE, Stern S, Andersson–Eklund L, Andersson L (1999) Confirmed quantitative trait loci for fatness and growth on pig chromosome 4. Heredity 82(Pt 2), 134–141
Milan D, Bidanel JP, Iannuccelli N, Riquet J, Amigues Y, et al. (2002) Detection of quantitative trait loci for carcass composition traits in pigs. Genet Sel Evol 34, 705–728
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16, 1215
Moller M, Berg F, Riquet J, Pomp D, Archibald A, et al. (2004) High-resolution comparative mapping of pig Chromosome 4, emphasizing the FAT1 region. Mamm Genome 15, 717–731
Ovilo C, Pérez–Enciso M, Barragan C, Clop A, Rodriguez C, et al. (2000) A QTL for intramuscular fat and back fat thickness is located on porcine chromosome 6. Mamm Genome 11, 344–346
Ovilo C, Clop A, Noguera JL, Oliver MA, Barragan C, et al. (2002) Quantitative trait locus mapping for meat quality traits in an Iberian × Landrace F2 pig population. J Anim Sci 80, 2801–2808
Paszek AA, Wilkie PJ, Flickinger GH, Rohrer GA, Alexander LJ, et al. (1999) Interval mapping of growth in divergent swine cross. Mamm Genome 10, 117–122
Pérez–Enciso M, Misztal I (2004) Qxpak: a versatile mixed model application for genetical genomics and QTL analyses. Bioinformatics 20, 2792–2798
Pérez–Enciso M, Clop A, Noguera JL, Ovilo C, Coll A, et al. (2000) A QTL on pig chromosome 4 affects fatty acid metabolism: evidence from an Iberian by Landrace intercross. J Anim Sci 78, 2525–2531
Rattink AP, De Koning DJ, Faivre M, Harlizius B, van Arendonk JA, et al. (2000) Fine mapping and imprinting analysis for fatness trait QTLs in pigs. Mamm Genome 11, 656–661
Serra X, Gil F, Pérez–Enciso M, Oliver MA, Vázquez JM, et al. (1998) A comparison of carcass, meat quality and histochemical characteristics of Iberian and Landrace pigs. Livest Prod Sci 56, 215–223
Turri MG, DeFries JC, Henderson ND, Flint J (2004) Multivariate analysis of quantitative trait loci influencing variation in anxiety-related behavior in laboratory mice. Mamm Genome 15, 69–76
Varona L, Ovilo C, Clop A, Noguera JL, Pérez–Enciso M, et al. (2002) QTL mapping for growth and carcass traits in an Iberian by Landrace pig intercross: additive, dominant and epistatic effects. Genet Res 80: 145-154
Varona L, Gomez–Raya L, Rauw WM, Clop A, Ovilo C, et al. (2004) Derivation of a Bayes factor to distinguish between linked or pleiotropic quantitative trait loci. Genetics 166, 1025–1035
Walling GA, Archibald AL, Cattermole JA, Downing AC, Finlayson HA, et al. (1998) Mapping of quantitative trait loci on porcine chromosome 4. Anim Genet 29, 415–424
Walling GA, Visscher PM, Andersson L, Rothschild MF, Wang L, et al. (2000) Combined analyses of data from quantitative trait loci mapping studies. Chromosome 4 effects on porcine growth and fatness. Genetics 155, 1369–1378
Wang L, Yu TP, Tuggle CK, Liu HC, Rothschild MF (1998) A directed search for quantitative trait loci on chromosomes 4 and 7 in pigs. J Anim Sci 76, 2560–2567
Wu R, Ma CX, Littell RC, Casella G (2002) A statistical model for the genetic origin of allometric scaling laws in biology. J Theor Biol 219, 121–135
Acknowledgments
This project was funded by Ministerio de Ciencia y Tecnologia (MCYT) (AGF99−0284-CO2) and (CPE03-010) grants. A. Mercadé was funded by a Formació Personal Investigador (FI) fellowship from the Generalitat de Catalunya, and J. Estellé was funded by a Formación Personal Universitario (FPU) grant from the Spanish Ministry of Education and Science (MEC). Part of the analyses was carried out in the Centre de Supercomputació de Catalunya (CESCA).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mercadé, A., Estellé, J., Noguera, J.L. et al. On growth, fatness, and form: A further look at porcine Chromosome 4 in an Iberian × Landrace cross. Mamm Genome 16, 374–382 (2005). https://doi.org/10.1007/s00335-004-2447-4
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s00335-004-2447-4