Abstract
Objectives
To establish a robust interpretable multiparametric deep learning (DL) model for automatic noninvasive grading of meningiomas along with segmentation.
Methods
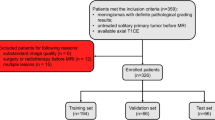
In total, 257 patients with pathologically confirmed meningiomas (162 low-grade, 95 high-grade) who underwent a preoperative brain MRI, including T2-weighted (T2) and contrast-enhanced T1-weighted images (T1C), were included in the institutional training set. A two-stage DL grading model was constructed for segmentation and classification based on multiparametric three-dimensional U-net and ResNet. The models were validated in the external validation set consisting of 61 patients with meningiomas (46 low-grade, 15 high-grade). Relevance-weighted Class Activation Mapping (RCAM) method was used to interpret the DL features contributing to the prediction of the DL grading model.
Results
On external validation, the combined T1C and T2 model showed a Dice coefficient of 0.910 in segmentation and the highest performance for meningioma grading compared to the T2 or T1C only models, with an area under the curve (AUC) of 0.770 (95% confidence interval: 0.644–0.895) and accuracy, sensitivity, and specificity of 72.1%, 73.3%, and 71.7%, respectively. The AUC and accuracy of the combined DL grading model were higher than those of the human readers (AUCs of 0.675–0.690 and accuracies of 65.6–68.9%, respectively). The RCAM of the DL grading model showed activated maps at the surface regions of meningiomas indicating that the model recognized the features at the tumor margin for grading.
Conclusions
An interpretable multiparametric DL model combining T1C and T2 can enable fully automatic grading of meningiomas along with segmentation.
Key Points
• The multiparametric DL model showed robustness in grading and segmentation on external validation.
• The diagnostic performance of the combined DL grading model was higher than that of the human readers.
• The RCAM interpreted that DL grading model recognized the meaningful features at the tumor margin for grading.




Similar content being viewed by others
Data availability
The data supporting this finding of this study are available from the corresponding author upon reasonable request.
Abbreviations
- 3D:
-
Three-dimensional
- AUC:
-
Area under the curve
- CI:
-
Confidence interval
- DL:
-
Deep learning
- IQR:
-
Interquartile range
- LRP:
-
Layer-wise relevance propagation
- RCAM:
-
Relevance-weighted Class Activation Mapping
- T1C:
-
Contrast-enhanced T1-weighted
- T2:
-
T2-weighted
- WHO:
-
World Health Organization
References
Ostrom QT, Gittleman H, Xu J et al (2016) CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2009–2013. Neuro Oncol 18:v1–v75
Kshettry VR, Ostrom QT, Kruchko C, Al-Mefty O, Barnett GH, Barnholtz-Sloan JS (2015) Descriptive epidemiology of World Health Organization grades II and III intracranial meningiomas in the United States. Neuro Oncol 17:1166–1173
Goldbrunner R, Minniti G, Preusser M et al (2016) EANO guidelines for the diagnosis and treatment of meningiomas. Lancet Oncol 17:e383-391
Modha A, Gutin PH (2005) Diagnosis and treatment of atypical and anaplastic meningiomas: a review. Neurosurgery 57:538-550; discussion 538-550
Huang RY, Bi WL, Weller M et al (2019) Proposed response assessment and endpoints for meningioma clinical trials: report from the Response Assessment in Neuro-Oncology Working Group. Neuro Oncol 21:26–36
Won SY, Park YW, Ahn SS et al (2021) Quality assessment of meningioma radiomics studies: bridging the gap between exploratory research and clinical applications. Eur J Radiol 138:109673
Nowosielski M, Galldiks N, Iglseder S et al (2017) Diagnostic challenges in meningioma. Neuro Oncol 19:1588–1598
Wadhwa A, Bhardwaj A, Singh Verma V (2019) A review on brain tumor segmentation of MRI images. Magn Reson Imaging 61:247–259
Harrison G, Kano H, Lunsford LD, Flickinger JC, Kondziolka D (2016) Quantitative tumor volumetric responses after Gamma Knife radiosurgery for meningiomas. J Neurosurg 124:146–154
Laukamp KR, Thiele F, Shakirin G et al (2019) Fully automated detection and segmentation of meningiomas using deep learning on routine multiparametric MRI. Eur Radiol 29:124–132
Laukamp KR, Pennig L, Thiele F et al (2020) Automated meningioma segmentation in multiparametric MRI : comparable effectiveness of a deep learning model and manual segmentation. Clin Neuroradiol. https://doi.org/10.1007/s00062-020-00884-4
Louis DN, Perry A, Reifenberger G et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131:803–820
Avants BB, Tustison N, Song G (2009) Advanced normalization tools (ANTS). Insight j 2:1–35
Roy S, Butman JA, Pham DL (2017) Robust skull stripping using multiple MR image contrasts insensitive to pathology. Neuroimage 146:132–147
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentationInternational Conference on Medical image computing and computer-assisted intervention. Springer, pp 234-241
Park YW, Jun Y, Lee Y et al (2021) Robust performance of deep learning for automatic detection and segmentation of brain metastases using three-dimensional black-blood and three-dimensional gradient echo imaging. Eur Radiol. https://doi.org/10.1007/s00330-021-07783-3
He K, Zhang X, Ren S, Sun J (2015) Delving deep into rectifiers: surpassing human-level performance on imagenet classificationProceedings of the IEEE international conference on computer vision, pp 1026-1034
Jun Y, Eo T, Kim T et al (2018) Deep-learned 3D black-blood imaging using automatic labelling technique and 3D convolutional neural networks for detecting metastatic brain tumors. Sci Rep 8:9450
Maas AL, Hannun AY, Ng AY (2013) Rectifier nonlinearities improve neural network acoustic modelsProc icml. Citeseer, pp 3
Ulyanov D, Vedaldi A, Lempitsky V (2016) Instance normalization: the missing ingredient for fast stylization. arXiv preprint arXiv:160708022
Paszke A, Gross S, Massa F et al (2019) Pytorch: an imperative style, high-performance deep learning library. arXiv preprint arXiv:191201703
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. arXiv preprint arXiv:14126980
Jeong Ryong Lee SK, Inyong Park, Taejoon Eo, Dosik Hwang (2021) Relevance-CAM: your model already knows where to lookIEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR)
Zeger SL, Liang KY, Albert PS (1988) Models for longitudinal data: a generalized estimating equation approach. Biometrics 44:1049–1060
Neromyliotis E, Kalamatianos T, Paschalis A et al (2020) Machine learning in meningioma MRI: past to present. A narrative review. J Magn Reson Imaging. https://doi.org/10.1002/jmri.27378
Ishi Y, Terasaka S, Yamaguchi S et al (2016) Reliability of the size evaluation method for meningiomas: maximum diameter, ABC/2 formula, and planimetry method. World Neurosurg 94:80–88
Laukamp KR, Pennig L, Thiele F et al (2020) Automated meningioma segmentation in multiparametric MRI. Clinical neuroradiology:1-10
Park YW, Shin SJ, Eom J et al (2022) Cycle-consistent adversarial networks improves generalizability of radiomics model in grading meningiomas on external validation. Sci Rep 12:7042
Zwanenburg A, Vallières M, Abdalah MA et al (2020) The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295:328–338
Moskowitz CS, Welch ML, Jacobs MA, Kurland BF, Simpson AL (2022) Radiomic analysis: study design, statistical analysis, and other bias mitigation strategies. Radiology 304:265–273
Banzato T, Causin F, Della Puppa A, Cester G, Mazzai L, Zotti A (2019) Accuracy of deep learning to differentiate the histopathological grading of meningiomas on MR images: a preliminary study. J Magn Reson Imaging 50:1152–1159
Boxerman JL, Quarles CC, Hu LS et al (2020) Consensus recommendations for a dynamic susceptibility contrast MRI protocol for use in high-grade gliomas. Neuro Oncol 22:1262–1275
Kaufmann TJ, Smits M, Boxerman J et al (2020) Consensus recommendations for a standardized brain tumor imaging protocol for clinical trials in brain metastases. Neuro Oncol 22:757–772
Paek SH, Kim SH, Chang KH et al (2005) Microcystic meningiomas: radiological characteristics of 16 cases. Acta Neurochir (Wien) 147:965-972; discussion 972
Verma PK, Nangarwal B, Verma J et al (2021) A clinico-pathological and neuro-radiological study of angiomatous meningioma: aggressive look with benign behaviour. J Clin Neurosci 83:43–48
Yao A, Pain M, Balchandani P, Shrivastava RK (2018) Can MRI predict meningioma consistency?: a correlation with tumor pathology and systematic review. Neurosurg Rev 41:745–753
Lin BJ, Chou KN, Kao HW et al (2014) Correlation between magnetic resonance imaging grading and pathological grading in meningioma. J Neurosurg 121:1201–1208
Kim JY, Park JE, Jo Y et al (2019) Incorporating diffusion- and perfusion-weighted MRI into a radiomics model improves diagnostic performance for pseudoprogression in glioblastoma patients. Neuro Oncol 21:404–414
Eo T, Jun Y, Kim T, Jang J, Lee HJ, Hwang D (2018) KIKI-net: cross-domain convolutional neural networks for reconstructing undersampled magnetic resonance images. Magn Reson Med 80:2188–2201
Jun Y, Shin H, Eo T, Kim T, Hwang D (2021) Deep model-based magnetic resonance parameter mapping network (DOPAMINE) for fast T1 mapping using variable flip angle method. Med Image Anal 70:102017
Choi KS, Sunwoo L (2022) Artificial intelligence in neuroimaging: clinical applications. Investigative Magnetic Resonance Imaging 26:1–9
Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A (2016) Learning deep features for discriminative localizationProceedings of the IEEE conference on computer vision and pattern recognition, pp 2921-2929
Bach S, Binder A, Montavon G, Klauschen F, Müller KR, Samek W (2015) On pixel-wise explanations for non-linear classifier decisions by layer-wise relevance propagation. PLoS One 10:e0130140
Shin H, Lee J, Eo T, Jun Y, Kim S, Hwang D (2020) The latest trends in attention mechanisms and their application in medical imaging. Taehan Yongsang Uihakhoe Chi 81:1305–1333
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D (2017) Grad-cam: visual explanations from deep networks via gradient-based localizationProceedings of the IEEE international conference on computer vision, pp 618-626
Park YW, Oh J, You SC et al (2019) Radiomics and machine learning may accurately predict the grade and histological subtype in meningiomas using conventional and diffusion tensor imaging. Eur Radiol 29:4068–4076
Park YW, Kim S, Ahn SS et al (2020) Magnetic resonance imaging-based 3-dimensional fractal dimension and lacunarity analyses may predict the meningioma grade. Eur Radiol 30:4615–4622
Kawahara Y, Nakada M, Hayashi Y et al (2012) Prediction of high-grade meningioma by preoperative MRI assessment. J Neurooncol 108:147–152
Shin HK, Park JH, Cho YH et al (2021) Risk factors for high-grade meningioma in brain and spine: systematic review and meta-analysis. World Neurosurg 151:e718–e730
Funding
Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, Information and Communication Technologies & Future Planning (2020R1A2C1003886); Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health & Welfare (HI21C1161); Basic Science Research Program through the NRF funded by the Ministry of Education (2020R1I1A1A01071648); Basic Science Research Program through the NRF funded by the Ministry of Science and ICT (2021R1A4A1031437, 2022R1A2C2008983); Brain Research Program through the NRF funded by the Ministry of Science, ICT & Future Planning (2018M3C7A1024734); Partially supported by the Yonsei Signature Research Cluster Program of 2022 (2022-22-0002); Artificial Intelligence Graduate School Program, Yonsei University [No. 2020-0-01361]; the KIST Institutional Program (Project No.2E31051-21-204).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Professor Seung-Koo Lee, MD, PhD, from Yonsei University College of Medicine (slee@yuhs.ac).
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors has significant statistical expertise (K.H, a biostatistics professor with 11 years of experience in biostatistics).
Informed consent
The institutional review board waived the requirement to obtain informed patient consent for this retrospective study.
Ethical approval
The Institutional Review Board waived the requirement of informed patient consent for this retrospective study.
Methodology
• retrospective
• diagnostic or prognostic study
• multicenter study
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jun, Y., Park, Y.W., Shin, H. et al. Intelligent noninvasive meningioma grading with a fully automatic segmentation using interpretable multiparametric deep learning. Eur Radiol 33, 6124–6133 (2023). https://doi.org/10.1007/s00330-023-09590-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-023-09590-4