Abstract
Objectives
Over the course of their treatment, patients often switch hospitals, requiring staff at the new hospital to import external imaging studies to their local database. In this study, the authors present MOdality Mapping and Orchestration (MOMO), a Deep Learning–based approach to automate this mapping process by combining metadata analysis and a neural network ensemble.
Methods
A set of 11,934 imaging series with existing anatomical labels was retrieved from the PACS database of the local hospital to train an ensemble of neural networks (DenseNet-161 and ResNet-152), which process radiological images and predict the type of study they belong to. We developed an algorithm that automatically extracts relevant metadata from imaging studies, regardless of their structure, and combines it with the neural network ensemble, forming a powerful classifier. A set of 843 anonymized external studies from 321 hospitals was hand-labeled to assess performance. We tested several variations of this algorithm.
Results
MOMO achieves 92.71% accuracy and 2.63% minor errors (at 99.29% predictive power) on the external study classification task, outperforming both a commercial product (82.86% accuracy, 1.36% minor errors, 96.20% predictive power) and a pure neural network ensemble (72.69% accuracy, 10.3% minor errors, 99.05% predictive power) performing the same task. We find that the highest performance is achieved by an algorithm that combines all information into one vote-based classifier.
Conclusion
Deep Learning combined with metadata matching is a promising and flexible approach for the automated classification of external DICOM studies for PACS archiving.
Key Points
• The algorithm can successfully identify 76 medical study types across seven modalities (CT, X-ray angiography, radiographs, MRI, PET (+CT/MRI), ultrasound, and mammograms).
• The algorithm outperforms a commercial product performing the same task by a significant margin (> 9% accuracy gain).
• The performance of the algorithm increases through the application of Deep Learning techniques.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Machine learning has had a large impact on the field of medicine, particularly in radiology [1]. Great strides have been made in the automated assessment of medical images in decision-making [2], prediction [3], and diagnostics [4]. An often-overlooked application of Deep Learning is the elimination of repetitive tasks in clinical routines. One of these tasks, currently requiring dedicated medical-technical personnel, is the processing and archiving of external DICOM studies. Frequently, patients from other facilities submit their imaging studies, which are then archived in the local hospital PACS. The facility of origin of these studies occasionally has a different language, a different naming standard for procedures, differently composed imaging studies, or even different procedures. Adherence to DICOM standards, such as clean labeling, file structure, and ordering (see for example [5]), is not always guaranteed, and Güld et al [6] have found that DICOM metadata is often unreliable (the DICOM tag Body Part Examined was found to be incorrect in 15.3% of cases). Studies that are thus incorrectly mapped to the local study nomenclature cause problems, from medical staff being unable to identify the relevant study for diagnostics and treatment, to arranging for a procedure to be performed unnecessarily. The lack of adherence to established standards additionally gives rise to a compositionality problem—a single study can contain multiple different series which together comprise one class, and such a composition can be incompatible with the classification scheme of the recipient hospital.
Related works have performed body region classification using various Deep Learning strategies (see [7,8,9,10,11]), but none with the explicit goal of study classification for PACS archiving in mind, nor for this comprehensive list of modalities.
In contrast to Dratsch et al [7], who used a neural network for evaluation of radiographs in the same context, the authors of this paper propose to extend the scope of the application to multiple modalities simultaneously—CT, X-ray angiography, radiographs, MRI, PET, ultrasound, and mammograms. Firstly, we aim to provide an automated classification algorithm, which can be integrated in the clinical routine. Secondly, we aim to establish the usefulness of neural networks in this context, as a part of or as a standalone solution. In this study, we propose a Deep Learning–based approach and compare its performance to a commercial product.
Materials and methods
Training images
For this retrospective study, we retrieved 11,934 de-identified imaging series belonging to 4597 separate examinations from the local PACS. These imaging series covered random timeframes and patients, and originated from 76 different types of studies. We acquired around 150–200 imaging series for each class, or fewer if unavailable. These classes (see Supplementary Materials) comprise the most common study types at our hospital. Imaging series were automatically labeled according to the class they were saved as in the PACS. Images were only rejected based on their series descriptor (excluding non-representative series such as topograms), but not quality or demographics. This is justified, as the external test set might feature low-quality scans and previously unseen image compositions. Ten percent (each) of the images are randomly drawn from the internal dataset to create a validation and test set for performance evaluation.
External studies
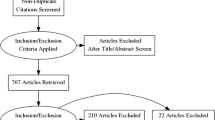
We collected 843 external studies (from 522 patients arriving at the local hospital from January 4, 2021, to January 8, 2021), which were labeled by a radiographer of the medical-technical staff with several years of experience as a team leader. Every study received at least one label, and some studies received several labels if they could be reasonably interpreted as multiple study types. This external test set was not used during training or for fine-tuning. Accuracy and predictive power on this set yield the final performance measures. A breakdown of the origins of the external studies can be found in Table 1.
In order to establish a measure of interrater reliability, another rater, a second-year resident in oncology with experience in evaluation of radiological images, labeled a subset of 300 of the external studies. The second rater was blinded to the labels of the first rater. Labels were considered to be in agreement if at least one of the labels provided by both raters matched.
The anonymization was performed using DicomDeidentify. A full list of DICOM tags that are erased or set to a common default value is provided by the authors in their code repository (https://github.com/luckfamousa/DicomDeidentify).
At our institution, imaging studies follow a specific structure (one study folder containing several series folders, each containing only the DICOM files of its respective series) and the DICOM tag entries, including the procedure names, are standardized. For external studies, this is often not the case, and neither standard is guaranteed. Thus, our acquisition of metadata is programmed to be agnostic to the structure of the imaging study, and we always evaluate several different tags.
Image preprocessing and neural network training
We evaluated two neural networks, a Resnet-152 [12] and DenseNet-161 [13], both pre-trained on ImageNet [14]. Images fed into the networks were first normalized into the range [0,1]. All image series were reduced to single-channel, greyscale images, and resampled to 512 × 512 along the X- and Y-axis, using spline interpolation. Two-dimensional image series became 512 × 512 × 3 images by stacking the first, middle, and last layer along the Z-axis. If only a single image was in the series (e.g., a single sonographic image), the same image was reused for all three channels. Three-dimensional images were resampled to length 512 along the Z-axis. The 512 × 512 × 3 input images were created by constructing the maximum intensity projections (MIPs) along the major axes. To avoid resampling errors, series where Z < 40 were treated as two-dimensional. Typical examples for two-dimensional images in this context are single sonographic images or a set of radiographic images from different perspectives. Typical examples for three-dimensional images are high-resolution CT or MRI image series. The process is depicted in Fig. 1.
If an image had a time axis, either it was treated as a two-dimensional image series (for 2D+T images), and the 512 × 512 × 3 image was created by stacking the first, middle, and last time step, or it was sampled at the halfway point in time and then treated like a regular three-dimensional image (for 3D+T images).
Images were randomly flipped or rotated during training (with probability p = 0.2), as image orientation can vary on external images. Performance was evaluated using cross-entropy loss. The networks were trained using transfer learning, freezing every layer except the final layer and the classifier (for hyperparameters, see Supplementary Materials). To eliminate unnecessary cross-contamination, we created an independent network for every modality except PET. For PET CT/MRI studies, the CT/MRI images were evaluated by the respective network. The PET images themselves were not used during training or inference.
Additionally, temperature scaling was performed using the hold-out validation set, so the network confidences become statistically meaningful probabilities [15]. This has a positive effect on study prediction accuracy (see Supplementary Materials).
MOMO
The MOdality Mapping and Orchestration’s (MOMO) algorithm follows a hierarchical layer structure (see Fig. 2). During each step, MOMO attempts a prediction. If successful, it returns the prediction, else it proceeds to the next step. In the default configuration, it begins by matching the Procedure Code (DICOM tag) of the study against known items. Next, the Study Description is evaluated. If both fail, it attempts to partially match the Study Description. After that, it extracts all pre-specified metadata keys from every series in the study and attempts to match these against a list of pre-specified keywords. Each match is weighted equally, with a simple majority choosing the prediction. A rules system disallows or modifies some votes based on a configuration file (see Supplementary Materials). In case of a tie, or if no votes were given, all imaging series are resampled and fed into the neural network corresponding to their modality. Each network prediction yields one vote, weighted by the confidence of the prediction. All votes are summed and evaluated.
MOMO structure. MOMO extracts meta- and imaging data and then progresses through its layers one by one until a decision is reached. The voting approach used for combining information is detailed in the “Methods” section. The entire algorithm can be modified from the Modality Mapping Database (a single tabular reference document containing all study classes and associated keywords), and a single configuration file (which holds technical parameters). This figure depicts one specific configuration of MOMO (the default configuration). It corresponds to the first algorithm in Table 2, labeled “NWE in layer 5,” as the network ensemble is the fifth resource to be used
We designed and evaluated a small Monte Carlo experiment as a proof of concept for this voting approach in an idealized setting (see Supplementary Materials).
The code, including instructions and a publicly available test case, is available from https://github.com/TIO-IKIM/MOMO_Submission_Code.
Statistical analysis
To assess the performance of the neural networks on a per-series basis, its accuracy is calculated for the internal test set, using fivefold cross-validation. Accuracies are reported with 95% confidence intervals.
MOMO is evaluated on the external dataset and compared to the Pan-Importer [16], a commercial product performing the classification task at our facility. An ablation study is performed, where the decision power of the network is increased by moving the network higher in the decision hierarchy (see Table 2). For every algorithm, we report the percentage of correct predictions, minor and major errors, accuracy, and network contribution, on the external test set. An error is considered minor if the classification is correct but too general (e.g., predicting “MR Spine” instead of “MR Thoracic Spine”), since this will not cause unnecessary examinations to be performed or studies not to be found. Additionally, we report which part of each algorithm was responsible for how many predictions.
A breakdown of the layer structure for the variations of our algorithm reported in the results section can be found in Table 2.
Results
We justified the MOMO voting approach (Fig. 1) in a Monte Carlo experiment (see Supplementary Material), showing that study-class prediction accuracy non-linearly increases with an increasing number of series in a study.
We evaluated multiple neural networks for different imaging modalities. The accuracies for the networks are shown in Table 3 and compared to the state of the art (SOTA), if available. We achieved comparable performance across all modalities. Conventional radiographs achieved the highest prediction accuracy (97.1%, CI: 96.2–98.0%), outperforming the current SOTA [7], and ultrasounds the lowest (81.4%, CI: 76.7–86.1%).
We also explored the study classification performance of MOMO, testing a wide range of settings and methods to combine metadata and image information. The results of the performance evaluation can be seen in Fig. 3. The network ensemble alone achieved 99.05% predictive power, 72.69% accuracy, and 10.3% minor errors. The commercial product reached 96.20% predictive power, 82.86% accuracy, and 1.36% minor errors.
Performance evaluation. A stacked bar plot, displaying correct predictions, minor and major errors for different study classifiers. Included are MOMO with and without network ensemble (NWE) with varying decision power (refer to Table 2 for details), as well as a classifier based purely on neural networks and the commercial product. Additionally, two hybrid variants, where the network votes together with other layers (merged votes), are tested. On the “Minimal Vote Rules” (MinVR) setting, fewer rules for vote modification (see the “MOMO” section) were applied for technical reasons. We report classification accuracy, number of correct, and incorrect predictions (split by severity of error) and the contribution of the networks (if any)
Overall, we found that a hybrid version of MOMO yielded the best performance, utilizing all metadata and neural networks in a single large vote. It outperformed other variants by a significant margin, scoring 99.29% predictive power, 92.71% accuracy, and 2.61% minor errors (using the networks with the best cross-validation performance).
Given the agreement criterion described in the “External studies” section, interrater agreement was 96.3% with a Cohen’s kappa of κ = 0.958. This value can be considered an approximate baseline for human performance on this task.
Figure 4 shows a breakdown of which information is responsible for the predictions for different variations of MOMO. The neural network ensemble has the highest prediction power (99.05% predictive power, 90.27% predictive power if exact matches for Procedure Code or Study Description are evaluated first), followed by a fuzzy matching of the Study Description (74.40% predictive power if exact matches are evaluated first).
Decision breakdown. A stacked bar plot, displaying which resource MOMO used to make a decision. The algorithms are ordered as in Fig. 3. The decisions are also sorted by their layer in the algorithm. For example, if the “Procedure Code Match” was checked first in an algorithm, it appears on the left end of the bar for that algorithm
Discussion
Different hospitals use different approaches to structure and name imaging studies. This generally necessitates manual identification of such studies before they are imported into local databases. In this study, we presented MOMO, a metadata-driven algorithm, supported by an ensemble of deep neural networks. It was trained to recognize body regions and automatically classify studies. We found that the network ensemble alone can perform similarly to a commercial product, while MOMO outperforms both. We found that the neural networks offered a boost in predictive power and the best performance in our task was achieved with a variant of MOMO utilizing neural networks.
The state of the art for comparable single series prediction is given by Dratsch et al [7] for plain radiographs and Raffy et al [8] for CT and MRI, while no Deep Learning body-region classification exists for ultrasound or X-ray angiography. Our training results compare favorably to the SOTA for radiographs. For CT and MRI, our accuracies are comparatively lower, but remain fairly competitive. There are multiple explanations for this. The training set in [8] covered a larger domain (multiple hospitals, scanner types, etc.) and contained more images, likely allowing for better generalization. Beyond that, as our labels come from the PACS and not manual labeling, some labels may either be false (an abdomen study also containing a series of the chest, which was subsequently labeled “abdomen”) or correct but difficult to identify (such as a vessel CT, which can have examples of various body sites in its corresponding studies). Since the latter problems are intrinsic to the compositionality of the study classification problem, one could also argue that the comparison is not valid.
We observed that the network ensemble offered an improvement over no-network variants of MOMO (see Fig. 3). However, this improvement only applied if the networks were not given too much decision power or if the network and metadata votes were combined for more robustness. The best performance was achieved by an algorithm which combined all information in such a manner. Finally, we note that, qualitatively, the erroneous predictions made by MOMO have an increased tendency to be anatomically related to the truth, compared to the commercial product (for a comparison of their relative rates of minor and major errors, see Fig. 3).
This work has limitations. If many similar keywords are added to the reference database of the algorithm, it can cause false predictions to increase due to false positives. Some classes are underrepresented during training, potentially worsening generalization. Additionally, the dataset used for training is automatically labeled, which may decrease the performance of the neural networks. Similarly, the compositionality of different types of study poses problems for neural networks that learn to predict based on single series. Finally, the origin (single institution) and amount of training data are limiting factors.
In the future, the use of additional training data (both local and external data) will allow the networks to mitigate some of these limitations. Additionally, new preprocessing steps (like contrast enhancement or cropping) could be introduced. Furthermore, improvements could be made by incorporating new decision layers into MOMO or improving the string matching (by comparing string similarity between metadata and keywords using metrics such as Levenshtein distance). Another interesting approach may be to perform predictions directly at the study level, using a Multiple Instance Learning classifier, which is particularly well-suited to solving the problem of compositionality we experienced when classifying DICOM studies. Besides this, future work could explore the theoretical viability of the voting approach in controlled settings with varying degrees of fuzzy labels. Finally, we posit that improved standardization of metainformation and reporting across institutions, while difficult to establish in practice, would be the most straightforward and promising approach to solving the challenge that MOMO was developed for.
In summary, this is the first open-source study classification tool to use metadata and neural networks for its decision process. It is fully automated and covers all common radiological modalities, offering increased quality compared to a commercial product performing the same task, as well as reduced workload for the clinical staff.
Abbreviations
- CR:
-
Conventional radiograph
- DICOM:
-
Digital Imaging and Communications in Medicine
- MG:
-
Mammogram
- MOMO:
-
Modality Mapping and Orchestration (the algorithm designed in this paper)
- PACS:
-
Picture Archiving and Communication Systems
- XA/DSA:
-
X-ray/digital subtraction angiography
References
European Society of Radiology (ESR) (2019) Impact of artificial intelligence on radiology: a EuroAIM survey among members of the European Society of Radiology. Insights Imaging 10:105
Wang JX, Sullivan DK, Wells AJ, Wells AC, Chen JH (2019) Neural networks for clinical order decision support. AMIA Jt Summits Transl Sci Proc 2019:315–324
Byun S-S, Heo TS, Choi JM et al (2021) Deep learning based prediction of prognosis in nonmetastatic clear cell renal cell carcinoma. Sci Rep 11:1242
Zaharchuk G (2020) Fellow in a box: combining AI and domain knowledge with bayesian networks for differential diagnosis in neuroimaging. Radiology 295:638–639
Bidgood WD, Horii SC, Prior FW, Van Syckle DE (1997) Understanding and using DICOM, the data interchange standard for biomedical imaging. J Am Med Inform Assoc 4:199–212
Gueld MO, Kohnen M, Keysers D et al (2002) Quality of DICOM header information for image categorization. In: SPIE Proceedings 4685:280–287
Dratsch T, Korenkov M, Zopfs D et al (2021) Practical applications of deep learning: classifying the most common categories of plain radiographs in a PACS using a neural network. Eur Radiol 31:1812–1818
Raffy P, Pambrun J-F, Kumar A et al (2021) Deep learning body region classification of MRI and CT examinations. https://doi.org/10.48550/arXiv.2104.13826
Zhang P, Wang F, Zheng Y (2017) Self supervised deep representation learning for fine-grained body part recognition. In: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017). IEEE, Melbourne, Australia, pp 578–582
Sugimori H (2018) Classification of computed tomography images in different slice positions using deep learning. J Healthc Eng 2018:1–9
Yan K, Lu L, Summers RM (2018) Unsupervised body part regression via spatially self-ordering convolutional neural networks. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). IEEE, Washington, DC, pp 1022–1025
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE, Las Vegas, NV, USA, pp 770–778
Huang G, Liu Z, van der Maaten L, Weinberger KQ (2017) densely connected convolutional networks. In: CVPR. pp 2261–2269
Deng J, Dong W, Socher R et al (2009) ImageNet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition. IEEE, Miami, FL, pp 248–255
Guo C, Pleiss G, Sun Y, Weinberger KQ (2017) On calibration of modern neural networks. In: Precup D, Teh YW (eds) Proceedings of the 34th International Conference on Machine Learning. PMLR, pp 1321–1330
Pansoma GmbH PAN Import, url: https://www.pansoma.at/produkte/pan-import/
Acknowledgements
The authors thank Jacob Murray for his contribution in proofreading the manuscript
Funding
Open Access funding enabled and organized by Projekt DEAL. Funding for this work was provided by the German State Ministry of Culture and Science NRW, for research under the Cancer Research Center Cologne Essen (CCCE) foundation. The funding was not provided specifically for this project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Prof. Jens Kleesiek.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board due to the retrospective and technical nature of the study.
Ethical approval
Institutional Review Board approval was obtained (Local IRB Application-Nr.: 20-9745-BO).
Methodology
• retrospective
• technical study
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 256 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jonske, F., Dederichs, M., Kim, MS. et al. Deep Learning–driven classification of external DICOM studies for PACS archiving. Eur Radiol 32, 8769–8776 (2022). https://doi.org/10.1007/s00330-022-08926-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-08926-w