Abstract
Purpose
The aim of this study was to develop and test a post-processing technique for detection and classification of lesions according to the BI-RADS atlas in automated breast ultrasound (ABUS) based on deep convolutional neural networks (dCNNs).
Methods and materials
In this retrospective study, 645 ABUS datasets from 113 patients were included; 55 patients had lesions classified as high malignancy probability. Lesions were categorized in BI-RADS 2 (no suspicion of malignancy), BI-RADS 3 (probability of malignancy < 3%), and BI-RADS 4/5 (probability of malignancy > 3%). A deep convolutional neural network was trained after data augmentation with images of lesions and normal breast tissue, and a sliding-window approach for lesion detection was implemented. The algorithm was applied to a test dataset containing 128 images and performance was compared with readings of 2 experienced radiologists.
Results
Results of calculations performed on single images showed accuracy of 79.7% and AUC of 0.91 [95% CI: 0.85–0.96] in categorization according to BI-RADS. Moderate agreement between dCNN and ground truth has been achieved (κ: 0.57 [95% CI: 0.50–0.64]) what is comparable with human readers. Analysis of whole dataset improved categorization accuracy to 90.9% and AUC of 0.91 [95% CI: 0.77–1.00], while achieving almost perfect agreement with ground truth (κ: 0.82 [95% CI: 0.69–0.95]), performing on par with human readers. Furthermore, the object localization technique allowed the detection of lesion position slice-wise.
Conclusions
Our results show that a dCNN can be trained to detect and distinguish lesions in ABUS according to the BI-RADS classification with similar accuracy as experienced radiologists.
Key Points
• A deep convolutional neural network (dCNN) was trained for classification of ABUS lesions according to the BI-RADS atlas.
• A sliding-window approach allows accurate automatic detection and classification of lesions in ABUS examinations.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Breast cancer is one of the most common causes of cancer death in females, and mortality rates are increasing worldwide [1]. In total, 2.3 million women were diagnosed with breast cancer in 2020 alone, causing 685,000 deaths worldwide [2]. The life-time probability to develop breast cancer is estimated to be 12.3% [3] and 1-, 3-, and 5-year survival rates are 92%, 75%, and 73% [4].
Current screening programs most commonly rely on mammography, which is known to reduce breast cancer–related mortality by up to 45% [5]. In spite of its cost-effectiveness, mammography screening exhibits some shortcomings reducing the effectiveness in quality-controlled organized screening programs and opportunistic breast cancer screening. Due to the need of breast compression in conventional mammography, many women decide not to undergo screening mammographies because of the fear of pain [6]. Moreover, dense breast tissue decreases the sensitivity of conventional mammography in cancer detection [7,8,9,10]. For women with dense breast tissue or women unwilling to undergo mammography, breast ultrasound may be a robust alternative modality, used as an adjunct to mammography or as an independent screening modality providing an increased accuracy for cancer detection [11, 12] but results are highly dependent on operators’ skill and experience, on top of that requiring an expert’s interpretation of results that may be highly subjective.
The Breast Imaging-Reporting and Data System (BI-RADS) was developed by the American College of Radiology (ACR) as a classification system to standardize quality control and risk assessment in mammography [13, 14]. The BI-RADS guidelines apply a numeric scale to describe the presence of lesions and the probability for malignancy. BI-RADS 1 corresponds to an image with no masses or lesions detected; BI-RADS 2 describes a lesion with no suspicion for malignancy; BI-RADS 3 stands for low probability of malignancy (< 3%); 4a—low suspicion of malignancy (2–10%); 4b—intermediate suspicion (11–50%); 4c—high suspicion (51–94%); and BI-RADS 5 corresponds to high probability of malignancy (> 95%) [15].
Automated breast ultrasound (ABUS) may be used instead of the more commonly applied hand-held ultrasound (HHUS) allowing a standardization of the image acquisition, complete coverage of the whole breast volume, and the delegation of the acquisition task to the technician [16, 17]. In first clinical studies, ABUS demonstrated similar accuracy as hand-held ultrasound; moreover, ABUS increased the cancer detection rate in combination with conventional mammography [18, 19]. One of the disadvantages of ABUS examinations is the large number of images resulting in an increased reading time for inexperienced radiologists. Additionally, a higher reader dependence of ABUS assessments has been reported [20].
Machine learning techniques have been shown to allow detection and classification of breast imaging findings [21], and particularly deep convolutional neural networks (dCNNs) provide a powerful tool in breast imaging, for example in detection and classification of microcalcifications in mammography [22], as well as the detection of breast cancer in hand-held US images and mammographies [23]. Applying dCNNs for feature extraction from ABUS examinations resulted in an increase of diagnostic reliability for second reading [24].
In this study, we trained a dCNN algorithm with ABUS findings to imitate the radiological decision-making according to the ACR BI-RADS recommendations of risk assessment in ABUS examinations. Moreover, a sliding-window approach was implemented for automatic localization of suspicious and non-suspicious lesions in ABUS datasets based on the dCNN classification.
Materials and methods
Patient data
This retrospective study has been approved by the local ethics committee (“Kantonale Ethikkommission Zurich”; Approval Number: 2016-00064). ABUS images for training, validation, and testing of the algorithm have been retrospectively gathered from the PACS database of the University Hospital of Zurich among the exams performed between 2017 and 2019. In total, 645 ABUS image sets containing lesions from 113 female patients were selected by an experienced radiologist with over 8 years of experience in breast imaging and over 3 years of experience in ABUS imaging. From the selected data, 55 patients had lesions of high probability for malignancy (BI-RADS 4/5) and 58 patients had lesions of low probability of malignancy (BI-RADS 2/3), all later confirmed as stable lesions or histologically proven fibroadenomas. For training of a model, 189 images containing a lesion with high malignancy probability and 178 images with low probability were used. For model validation, each BI-RADS category contained 75 images. Dataset for testing comprised of 71 images with high malignancy probability from 10 patients and 57 images with low malignancy probability from 10 patients that were not used for training nor validation of a model.
ABUS examination
Images were acquired with a dedicated ABUS device (Invenia™ Automated Breast Ultrasound System, GE Healthcare) by a trained technician. Acquisition consisted of three volumes per breast in order to cover the whole breast volume. Each image set consists of a stack of 2D images with a matrix size of 600 × 600 pixels; each of them presents a US view plane 0.5 mm apart from each other.
Image selection and data preparation
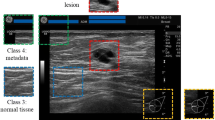
From the complete datasets, single images depicting lesions were selected by the radiologist and classified according to BI-RADS categories. Depiction of a single image from the image stack is shown in Fig. 1. Ten image sets were excluded from further analysis due to presence of artifacts. For training and validation of the dCNN, the area surrounding the lesion was selected by the radiologist and cropped as a rectangular image with a size of 151 × 181 pixels. Training/validation images were augmented to improve the generalization of training and to balance the classes of the dataset using the TensorFlow (Google LLC) data augmentation tool for rotation, height and width shift, shear, zoom, and horizontal flipping. Detailed data on the number of augmented images used for each part of learning process are presented in a Table 1. To allow for a step-wise detection and classification, 3 dCNN models were trained:
-
Model 1—distinguishing between image background from breast tissue
-
Model 2—distinguishing between normal breast tissue and presence of a lesion (both benign and malignant)
-
Model 3—determination of the BI-RADS category of the detected lesion (BI-RADS 2/3 or BI-RADS 4/5)
dCNN architecture
The models were trained using a deep convolutional neural network with 13 convolutional layers with 5 max pooling layers for downsampling followed by 2 dense layers with the ReLU activation function. A 50% dropout was applied to reduce overfitting and a softmax activation function was used for the final weights. For model training, a stochastic gradient descent optimization was used with binary cross-entropy loss function. Batches of 16 were applied and the learning rate was set to 1.0 × 10−6, a Nesterov momentum of 0.90 was used, and training was stopped after training and validation accuracies stopped improving, what meant 120 epochs for models 1 and 2 and 160 epochs for model 3. dCNNs were trained with a TensorFlow 2.0 software library on a machine running Ubuntu 16.04 OS with an Nvidia 1080 GTX GPU. Class weighting was used to reduce the imbalance in training datasets.
Image analysis and lesion detection
To localize regions of an image, in which tissue or lesions are depicted, and to create a probability map showing pixel-by-pixel the probability of lesion presence, a sliding-window approach was implemented in the programming language Python 3.5. From the original ABUS image with a size of 600 × 600 pixels, sections of 151 × 181 pixels around given x,y coordinates were cropped in a loop over x and y, and with each cropped image the step-wise dCNN classification algorithm was applied. Probability results of calculations were written into 3 arrays (results for each model is written into separate array) with corresponding x and y coordinates. After all the dCNN evaluations for one cropped area were done, the center on the original image was moved by 10 pixels in X or Y direction in the loop. A detailed scheme of the sliding-window approach is presented in Fig. 2.
Sliding-window approach for localization and prediction of lesions on images. For analysis of the whole image, separate crops are being selected (a). Prediction based on cropped image with a window of a fixed size (b). All 3 models are being applied to cropped fragment of an image. Probability result from each of them is written into table with corresponding coordinates of center of an image (c)
For each analyzed image, the sliding-window algorithm results in three 60 × 60 matrices consisting of the class probabilities of the step-wise classification. Results of calculations performed with 3 trained models are visualized as heatmaps giving spatial information on the probability of detection and classification. A visualization of an exemplary calculation is presented in Fig. 3. In the colormaps, results are presented as overlays on the original image in red and blue color representing the probabilities of dCNN classification to the BI-RADS 2/3 or BI-RADS 4/5 categories. Due to the step-wise classification of the image region, only those areas representing a detected lesion (evaluation step 2) were evaluated in step 3 using the model classifying into BI-RADS 2/3 or BI-RADS 4/5 (evaluation step 3). Therefore, only those areas which represent a detected lesion exhibit colors of red or blue in the colormap. To generate probability maps from the complete image volume, the previously described method was consecutively applied to all images from the image stack resulting in a matrix of 60 × 60 × 3 × 330 (3 probability matrices with a dimension 60 × 60 for each of the 330 images from a full examination set).
Probability maps are generated for each image. Sliding-window algorithm was applied to the original image (a). As a result of model 1 calculations, grayscale probability map was generated presenting area of breast tissue depiction (b). Application of model 2 on an image results in depiction of suspected lesion visualized by grayscale probability map (c). Lesions were classified according to BI-RADS with model 3 and visualized in blue (BI-RADS 2/3) and red (BI-RADS 4/5) colors (d)
Comparison with human reading
The test dataset was categorized into the three categories BI-RADS 2 (no suspicion of breast cancer), BI-RADS 3 (low probability of breast cancer), and BIRADS 4/5 (high likelihood of breast cancer) by two radiologists, reader 1 with more than 10 years of experience in breast imaging and reader 2 with more than 5 years of experience in breast imaging. All lesions were also classified according to the previously described 3 categories by the step-wise dCNN algorithm for lesion detection and classification.
Intra-reader reliability was obtained by a repeated categorization of the same datasets by the same radiologists after a period of 3 months (Fig. 4).
Statistical analysis
The statistical analysis was performed using the Scikit-learn 0.22.1 package for the Python programming language. Confusion matrices were applied to compare the performance of the dCNN with the ground truth categorization (human readers). Agreement of categorization between dCNN and human readers and inter- and intra-reader correlations have been calculated using Cohen’s kappa coefficient (κ). The diagnostic performances of the trained models were calculated using receiver operating characteristics. Diagnostic accuracy was evaluated in a form of binary classification accuracy and area under the curve (AUC) statistic of ROC curves with a confidence interval CI = 0.95 from Monte Carlo simulation.
Results
Model training
Training of the dCNN models resulted in accuracies between 98.2 and 99.0% on the training datasets and between 96.9 and 99.1% accuracy on the validation datasets. Training and validation curves for the model distinguishing between normal tissue and lesions (model 2) and for the model classifying a detected lesion in probably benign (BI-RADS 2/3) or suspicious (BI-RADS 4/5) are presented in Fig. 5. Detailed results of the training and validation accuracies and the number of images are presented in Table 1.
Validation on single images presenting lesions
For testing the dCNN models with data not used for training or validation, a dataset of 128 images was applied, each depicting a lesion of either BI-RADS 2/3 or BI-RADS 4/5, using the double-read radiological report from the RIS archive as ground truth. In the image analysis, the dCNN and two experienced human readers were compared treating each image as an independent case.
Using model 3 for analysis, an accuracy of 79.7% was found, and a ROC analysis for model 3 leads to an AUC of 0.91 (AUC: 0.91 [95% CI: 0.85–0.96]) with Youden’s index J of 0.92. AUC for reader 1 was 0.80 [95% CI: 0.73–0.87], and for reader 2 AUC was 0.71 [95% CI: 0.63–0.78]. Confusion matrices for the dCNN model and both human readers are presented in Table 2, detailed results regarding accuracies, inter-reader agreement, and ROC analyses are presented in Table 3 and Table 4, and corresponding ROC curves are depicted in Fig. 6.
The agreement between the dCNN model 3 and the ground truth was moderate (κ: 0.57 [95% CI: 0.50–0.64]). The agreement between reader 1 and the ground truth was substantial (κ: 0.61 [95% CI: 0.53–0.68]), whereas the agreement between reader 2 and the ground truth was moderate (κ: 0.44 [95% CI: 0.36–0.51]). Agreement between readers was moderate (κ: 0.43 [95% CI: 0.28–0.58]).
Intra-reader agreement was substantial for both reader 1 (κ: 0.64 [95% CI: 0.54–0.77]) and reader 2 (κ: 0.66 [95% CI: 0.54–0.78]).
Validation on full 3D ABUS datasets
Analyzing complete 3D ABUS datasets and classifying this dataset according to the highest BI-RADS score of lesions present, the accuracy of classification of the algorithm improved to 90.9% compared to the radiological report as a ground truth. ROC analyses for dCNN resulted in an AUC of 0.91 (AUC: 0.91 [95% CI: 0.77–1.00]) with Youden’s index J of 0.92. The AUC for reader 1 was 0.82 [95% CI: 0.68–1.00] and for reader 2 was 0.91 [95% CI: 0.77–1.00]. ROC curves for the analysis of complete ABUS datasets are depicted in Fig. 6, and the confusion matrices are shown in Table 5.
The agreement between the dCNN algorithm and the ground truth was almost perfect (κ: 0.82 [95% CI: 0.60–0.95]) and the agreement between readers and ground truth was substantial to almost perfect (reader 1—κ: 0.63 [95% CI: 0.47–0.81] and reader 2—κ: 0.82 [95% CI: 0.60–0.95]). Agreement between readers was moderate (κ: 0.46 [95% CI: 0.09–0.82]).
Intra-reader agreement was fair (κ: 0.32 [95% CI: 0.01–0.65]) for reader 1 and almost perfect (κ: 0.81 [95% CI: 0.56–1.00]) for reader 2.
Discussion
In the current study, we were able to show that a deep convolutional neural network (dCNN) can be trained to classify lesions in ABUS according to the ACR BI-RADS classification with accuracies higher than 95% in both the training and the validation datasets. Compared to human reading by experienced radiologists, the dCNN showed similar diagnostic accuracy on test datasets not used for training of the machine learning algorithm, and also inter-reader agreement between the dCNN was comparable to the inter-reader agreement of experienced radiologists. Applying a 3-dimensional sliding-window approach with a step-wise assessment of the region of interest, the dCNN was able to detect and classify lesions according to the BI-RADS atlas with accuracy providing a technique for the automatic reading of 3D ABUS datasets mimicking the human workflow.
In the clinical routine, the evaluation of radiological breast images is standardized according to the BI-RADS atlas grading the probability of the presence of breast cancer based on atlas images. In spite of this standardization, the quality of radiological decision-making is dependent on the experience of the reader as well as the workload. It has been shown that even after short-term re-evaluation of images, up to 29% of BI-RADS assessments may be reclassified [25]. The usability of AI models as a support tool for radiologists in the decision-making process has been shown in several studies, e.g. [26], which particularly can be used to decrease the false-positive rates in mammography screening [27].
Ultrasound as an adjunct to mammography has been shown to decrease the false-negative rate of conventional mammography, and ABUS may be used instead of hand-held ultrasound exhibiting similar cancer detection rates [28]. As the ABUS examination is typically carried out by the technician, a workflow improvement is achieved in radiological institutions replacing hand-held ultrasound with ABUS. Moreover, as the complete breast volume is examined and stored as 3D datasets, ABUS is a step further in the standardization of breast imaging workflow. Another unwanted effect of ABUS is the relatively long reading time and the higher rate of false-negative lesions. Here, our proposed algorithm may offer a significant improvement: it highlights suspicious lesions in the large volumetric 3D dataset and gives also a suggestion of the correct classification of the lesion, which may be helpful for inexperienced readers. Application of AI in clinical routine potentially offers a reliable second opinion tool for experienced radiologists as well as providing a training platform for inexperienced readers. An AI solution is observer-independent and minimizes the impact of experience and external factors for diagnosis. Instead of working as a “black-box,” the explainable AI concept is applied in this study. By providing step-wise solutions, each decision made by the AI program can be traced back, which enables deeper understanding and adaptation of the AI program in clinical use.
The accuracy of our algorithm trained for classification of ABUS lesions is comparable to the results of Wang et al [24], who used multiview CNNs to classify breast lesions in datasets obtained from a breast volume scanner with accuracy in the order of 88%. Lee et al showed that similar algorithms allow for precise lesion segmentation with Spearman’s correlation coefficient of ρ = 0.929 [29].
In clinical usage, CAD software has proven to significantly improve and accelerate workflow. Those systems have already proven its high sensitivity in mammography with detection of masses of 91% on initial and 89% on follow-up mammograms [30] as well as ultrasonography with sensitivity of 90.9% [31]. ABUS’s main advantages over other examination types, like volumetric representation and relative procedure simplicity, increase requirements for results analysis for radiologists. CAD systems allow to overcome this issue and provide reliable second opinion to specialist [32]. The application of CAD software for ABUS in clinical use demonstrated an evaluation speed up by 9–15% [33, 34]. Segmentation methods used for CAD have shown to improve the ability to distinguish positive from negative cases (AUC) by 6% when used as support program by inexperienced readers [35]. Commercially available AI CAD software based on minimum intensity projection visualization (MinIP) improves sensitivity of the readout by 5.2–10.6%, but may come with a possible decrease of specificity in the range of 1.4–5.7%, while additionally requiring parameter adjustment for readout that affects sensitivity [36]. Even though proven to work in a clinical environment, those methods come with limitations in regard to irregularities and various lesion sizes what may lead to an increased number of false-positives [37] and may require parameter fine-tuning and extensive image preprocessing [38]. The presented method provides improved generalization and allows a possibility for customization for specific manufacturers and adjustment to internal work processes.
Our study has some limitations: (1) A retrospective study design was chosen for validating the algorithm resulting in potential bias due to the choice of datasets. However, a prospective evaluation was out of the scope of this methodological study. (2) All data was gathered from one single institution and data from only one device supplier was used for acquisition. Nonetheless, the purpose of this study was not to demonstrate that our algorithm works well on every possible clinical setup but to demonstrate in principle that an algorithm can be trained to mimic human reading in ABUS examinations. (3) Although data was derived from the general female patient cohort of our institution, not all ethnicities were included in this study. (4) Only one dCNN architecture and detection algorithm has been investigated and no comparison with pretrained models has been made; therefore, we cannot exclude that other dCNN architectures potentially would result in even higher accuracies.
In conclusion, the trained AI models allowed for further standardization as an independent second opinion tool in ABUS examinations according to BI-RADS catalogue. Implementation of our AI models into the clinical routine potentially could improve reliability of assessment while reducing workload for radiologists.
Abbreviations
- ABUS:
-
Automated breast ultrasound
- ACR:
-
American College of Radiology
- AUC:
-
Area under the curve
- BI-RADS:
-
Breast Imaging-Reporting and Data System
- CAD:
-
Computer-aided detection
- dCNN:
-
Deep convolutional neural network
- HHUS:
-
Hand-held ultrasound
- ROC:
-
Receiver operating characteristic
- US:
-
Ultrasound
References
Azamjah N, Soltan-Zadeh Y, Zayeri F (2019) Global trend of breast cancer mortality rate: a 25-year study. Asian Pac J Cancer Prev 20:2015–2020
World Health Organization (2021) Breast cancer. World Health Organization, Geneva. Available via https://www.who.int/news-room/fact-sheets/detail/breast-cancer
Rojas K, Stuckey A (2016) Breast cancer epidemiology and risk factors. Clin Obstet Gynecol. 59:651–672
Maajani K, Jalali A, Alipour S, Khodadost M, Tohidinik HR, Yazdani K (2019) The global and regional survival rate of women with breast cancer: a systematic review and meta-analysis. Clin Breast Cancer 19:165–177
Swedish Organised Service Screening Evaluation Group (2006) Reduction in breast cancer mortality from the organised service screening with mammography: 2. Validation with alternative analytic methods. Cancer Epidemiol Biomark Prev 15:52–56
Hofvind S, Geller BM, Skelly J, Vacek PM (2012) Sensitivity and specificity of mammographic screening as practised in Vermont and Norway. Br J Radiol 85:1226–1232
Lynge E, Vejborg I, Andersen Z, von Euler-Chelpin M, Napolitano G (2019) Mammographic density and screening sensitivity, breast cancer incidence and associated risk factors in Danish breast cancer screening. J Clin Med 8:2021
Boyd NF, Guo H, Martin LJ et al (2007) Mammographic density and the risk and detection of breast cancer. N Engl J Med 356:227–236
Buist DS, Porter PL, Lehman C, Taplin SH, White E (2004) Factors contributing to mammography failure in women aged 40-49 years. J Natl Cancer Inst 96:1432–1440
Boyd NF, Martin LJ, Bronskill M, Yaffe MJ, Duric N, Minkin S (2010) Breast tissue composition and susceptibility to breast cancer. J Natl Cancer Inst 102:1224–1237
Berg WA, Zhang Z, Lehrer D et al (2012) Detection of breast cancer with addition of annual screening ultrasound or a single screening MRI to mammography in women with elevated breast cancer risk. JAMA 307:1394–1404
Shin HJ, Kim HH, Cha JH (2015) Current status of automated breast ultrasonography. Ultrasonography 34:165–172
Liberman L, Menell JH (2002) Breast imaging reporting and data system (BI-RADS). Radiol Clin North Am 40:409–430
D’Orsi CJ, Bassett LW, Berg WA et al (2003) Breast Imaging Reporting and Data System. American College of Radiology
Magny SJ, Shikhman R, Keppke AL (2020) Breast Imaging Reporting and Data System. StatPearls Publishing
Abd Elkhalek YI, Bassiouny AM, Hamid RWARA (2019) Automated breast ultrasound system (ABUS): can it replace mammography as a screening tool? Egypt J Radiol Nucl Med 50:51
Wilczek B, Wilczek HE, Rasouliyan L, Leifland K (2016) Adding 3D automated breast ultrasound to mammography screening in women with heterogeneously and extremely dense breasts: report from a hospital-based, high-volume, single-center breast cancer screening program. Eur J Radiol 85:1554–1563
Tagliafico AS, Calabrese M, Mariscotti G et al (2016) Adjunct screening with tomosynthesis or ultrasound in women with mammography-negative dense breasts: interim report of a prospective comparative trial. J Clin Oncol 34:1882–1888
Mostafa AAE, Eltomey MA, Elaggan AM (2019) Automated breast ultrasound (ABUS) as a screening tool: initial experience. Egyptian Journal of Radiology and Nuclear Medicine 50:37
Skaane P, Gullien R, Eben EB, Sandhaug M, Schulz-Wendtland R, Stoeblen F (2015) Interpretation of automated breast ultrasound (ABUS) with and without knowledge of mammography: a reader performance study. Acta Radiol. 56:404–412
Marcon M, Ciritsis A, Rossi C et al (2019) Diagnostic performance of machine learning applied to texture analysis-derived features for breast lesion characterisation at automated breast ultrasound: a pilot study. Eur Radiol Exp 3:44
Schönenberger C, Hejduk P, Ciritsis A, Marcon M, Rossi C, Boss A (2020) Classification of mammographic breast microcalcifications using a deep convolutional neural network: a BI-RADS-based approach. Invest Radiol 56:224–231
Becker AS, Marcon M, Ghafoor S, Wurnig MC, Frauenfelder T, Boss A (2017) Deep learning in mammography: diagnostic accuracy of a multipurpose image analysis software in the detection of breast cancer. Invest Radiol 52:434–440
Wang Y, Choi EJ, Choi Y, Zhang H, Jin GY, Ko SB (2020) Breast cancer classification in automated breast ultrasound using multiview convolutional neural network with transfer learning. Ultrasound Med Biol 46:1119–1132
Kim WH, Moon WK, Kim SM et al (2013) Variability of breast density assessment in short-term reimaging with digital mammography. Eur J Radiol 82:1724–1730
Geras KJ, Mann RM, Moy L (2019) Artificial intelligence for mammography and digital breast tomosynthesis: current concepts and future perspectives. Radiology 293:246–259
Le EPV, Wang Y, Huang Y, Hickman S, Gilbert FJ (2019) Artificial intelligence in breast imaging. Clin Radiol 74:357–366
Giger ML, Inciardi MF, Edwards A et al (2016) Automated breast ultrasound in breast cancer screening of women with dense breasts: reader study of mammography-negative and mammography-positive cancers. AJR Am J Roentgenol. 206:1341–1350
Lee CY, Chang TF, Chou YH, Yang KC (2020) Fully automated lesion segmentation and visualization in automated whole breast ultrasound (ABUS) images. Quant Imaging Med Surg 10:568–584
Kim SJ, Moon WK, Cho N, Cha JH, Kim SM, Im JG (2008) Computer-aided detection in full-field digital mammography: sensitivity and reproducibility in serial examinations. Radiology 246:71–80
Tanaka H, Chiu SW, Watanabe T, Kaoku S, Yamaguchi T (2019) Computer-aided diagnosis system for breast ultrasound images using deep learning. Phys Med Biol 64:235013
Kozegar E, Soryani M, Behnam H, Salamati M, Tan T (2020) Computer aided detection in automated 3-D breast ultrasound images: a survey. Artif Intell Rev 53:1919–1941
Xu X, Bao L, Tan Y, Zhu L, Kong F, Wang W (2018) 1000-case reader study of radiologists’ performance in interpretation of automated breast volume scanner images with a computer-aided detection system. Ultrasound Med Biol 44:1694–1702
van Zelst JCM, Tan T, Clauser P et al (2018) Dedicated computer-aided detection software for automated 3D breast ultrasound; an efficient tool for the radiologist in supplemental screening of women with dense breasts. Eur Radiol 28:2996–3006
Tan T, Platel B, Twellmann T et al (2013) Evaluation of the effect of computer-aided classification of benign and malignant lesions on reader performance in automated three-dimensional breast ultrasound. Acad Radiol 20:1381–1388
van Zelst JCM, Tan T, Platel B et al (2017) Improved cancer detection in automated breast ultrasound by radiologists using Computer Aided Detection. Eur J Radiol 89:54–59
van Zelst JC, Tan T, Mann RM, Karssemeijer N (2020) Validation of radiologists’ findings by computer-aided detection (CAD) software in breast cancer detection with automated 3D breast ultrasound: a concept study in implementation of artificial intelligence software. Acta Radiol 61:312–320
Gardezi J (2017) Handbook of research on machine learning innovations and trends. IGI Global
Acknowledgements
This study has been supported by the Clinical Research Priority Program Artificial Intelligence in oncological Imaging of the University Zurich.
Funding
Open access funding provided by University of Zurich. The authors state that this work has not received any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Andreas Boss, MD PhD.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was obtained from all subjects (patients) in this study.
Ethical approval
Institutional review board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hejduk, P., Marcon, M., Unkelbach, J. et al. Fully automatic classification of automated breast ultrasound (ABUS) imaging according to BI-RADS using a deep convolutional neural network. Eur Radiol 32, 4868–4878 (2022). https://doi.org/10.1007/s00330-022-08558-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-08558-0