Abstract
Objective
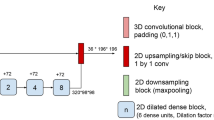
Automated quantification of infratentorial multiple sclerosis lesions on magnetic resonance imaging is clinically relevant but challenging. To overcome some of these problems, we propose a fully automated lesion segmentation algorithm using 3D convolutional neural networks (CNNs).
Methods
The CNN was trained on a FLAIR image alone or on FLAIR and T1-weighted images from 1809 patients acquired on 156 different scanners. An additional training using an extra class for infratentorial lesions was implemented. Three experienced raters manually annotated three datasets from 123 MS patients from different scanners.
Results
The inter-rater sensitivity (SEN) was 80% for supratentorial lesions but only 62% for infratentorial lesions. There was no statistically significant difference between the inter-rater SEN and the SEN of the CNN with respect to the raters. For supratentorial lesions, the CNN featured an intra-rater intra-scanner SEN of 0.97 (R1 = 0.90, R2 = 0.84) and for infratentorial lesion a SEN of 0.93 (R1 = 0.61, R2 = 0.73).
Conclusion
The performance of the CNN improved significantly for infratentorial lesions when specifically trained on infratentorial lesions using a T1 image as an additional input and matches the detection performance of experienced raters. Furthermore, for infratentorial lesions the CNN was more robust against repeated scans than experienced raters.
Key Points
• A 3D convolutional neural network was trained on MRI data from 1809 patients (156 different scanners) for the quantification of supratentorial and infratentorial multiple sclerosis lesions.
• Inter-rater variability was higher for infratentorial lesions than for supratentorial lesions. The performance of the 3D convolutional neural network (CNN) improved significantly for infratentorial lesions when specifically trained on infratentorial lesions using a T1 image as an additional input.
• The detection performance of the CNN matches the detection performance of experienced raters.





Similar content being viewed by others
Abbreviations
- BL:
-
Baseline
- CM:
-
Comparison map
- CNN:
-
Convolutional neural networks
- CSF:
-
Cerebrospinal fluid
- FLAIR:
-
Fluid-attenuated inversion recovery sequence
- FN:
-
False negative
- FP:
-
False positive
- FPR:
-
False positive rate
- FU:
-
Follow-up
- GT:
-
Ground truth
- Infra:
-
Infratentorial
- MRI:
-
Magnetic resonance imaging
- MS:
-
Multiple sclerosis
- MTA:
-
Medical technology associates
- PD:
-
Proton density (weighted MRI sequence)
- PPV:
-
Positive predictive value
- R1/R2:
-
Rater 1/rater 2
- SEN:
-
Sensitivity
- supra:
-
Supratentorial
- TP:
-
True positive
- VAL1/2/3:
-
Validation set 1/2/3
References
Polman CH, Reingold SC, Banwell B et al (2011) Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol 69:292–302
Rovira À, Wattjes MP, Tintoré M et al (2015) Evidence-based guidelines: MAGNIMS consensus guidelines on the use of MRI in multiple sclerosis-clinical implementation in the diagnostic process. Nat Rev Neurol 11:471–482
Rahn AC, Köpke S, Stellmann JP et al (2019) Magnetic resonance imaging as a prognostic disability marker in clinically isolated syndrome: a systematic review. Acta Neurol Scand 139:18–32
Minneboo A, Barkhof F, Polman CH, Uitdehaag BM, Knol DL, Castelijns JA (2004) Infratentorial lesions predict long-term disability in patients with initial findings suggestive of multiple sclerosis. Arch Neurol 61:217–221
Vellinga MM, Geurts JJ, Rostrup E et al (2009) Clinical correlations of brain lesion distribution in multiple sclerosis. J Magn Reson Imaging 29:768–773
Meier DS, Guttmann CRG, Tummala S et al (2018) Dual-sensitivity multiple sclerosis lesion and CSF segmentation for multichannel 3T brain MRI. J Neuroimaging 28:36–47
Filippi M, Preziosa P, Banwell BL et al (2019) Assessment of lesions on magnetic resonance imaging in multiple sclerosis: practical guidelines. Brain 142:1858–1875
Bailey WM (2007) Fast fluid attenuated inversion recovery (FLAIR) imaging and associated artefacts in magnetic resonance imaging (MRI). Radiography 13:283–290
Cabezas M, Oliver A, Roura E et al (2014) Automatic multiple sclerosis lesion detection in brain MRI by FLAIR thresholding. Comput Methods Programs Biomed 115:147–161
Griffanti L, Zamboni G, Khan A et al (2016) BIANCA (Brain Intensity AbNormality Classification Algorithm): a new tool for automated segmentation of white matter hyperintensities. Neuroimage 141:191–205
Roura E, Oliver A, Cabezas M et al (2015) A toolbox for multiple sclerosis lesion segmentation. Neuroradiology 57:1031–1043
Salem M, Cabezas M, Valverde S et al (2018) A supervised framework with intensity subtraction and deformation field features for the detection of new T2-w lesions in multiple sclerosis. Neuroimage Clin 17:607–615
Schmidt P, Gaser C, Arsic M et al (2012) An automated tool for detection of FLAIR-hyperintense white-matter lesions in multiple sclerosis. Neuroimage 59:3774–3783
Schmidt P, Pongratz V, Küster P et al (2019) Automated segmentation of changes in FLAIR-hyperintense white matter lesions in multiple sclerosis on serial magnetic resonance imaging. Neuroimage Clin 23:101849
Bernal J, Kushibar K, Asfaw DS et al (2019) Deep convolutional neural networks for brain image analysis on magnetic resonance imaging: a review. Artif Intell Med 95:64–81
Danelakis A, Theoharis T, Verganelakis DA (2018) Survey of automated multiple sclerosis lesion segmentation techniques on magnetic resonance imaging. Comput Med Imaging Graph 70:83–100
Krüger J, Opfer R, Gessert N et al (2020) Fully automated longitudinal segmentation of new or enlarged multiple sclerosis lesions using 3D convolutional neural networks. Neuroimage Clin 28:102445
La Rosa F, Abdulkadir A, Fartaria MJ et al (2020) Multiple sclerosis cortical and WM lesion segmentation at 3T MRI: a deep learning method based on FLAIR and MP2RAGE. Neuroimage Clin 27:102335
Manjón JV, Coupé P, Raniga P et al (2018) MRI white matter lesion segmentation using an ensemble of neural networks and overcomplete patch-based voting. Comput Med Imaging Graph 69:43–51
Valverde S, Salem M, Cabezas M et al (2019) One-shot domain adaptation in multiple sclerosis lesion segmentation using convolutional neural networks. Neuroimage Clin 21:101638
Biberacher V, Schmidt P, Keshavan A et al (2016) Intra- and interscanner variability of magnetic resonance imaging based volumetry in multiple sclerosis. Neuroimage. https://doi.org/10.1016/j.neuroimage.2016.07.035
Grahl S, Pongratz V, Schmidt P et al (2019) Evidence for a white matter lesion size threshold to support the diagnosis of relapsing remitting multiple sclerosis. Mult Scler Relat Disord 29:124–129
Opfer R, Suppa P, Kepp T, Spies L, Schippling S, Huppertz HJ (2016) Atlas based brain volumetry: how to distinguish regional volume changes due to biological or physiological effects from inherent noise of the methodology. Magn Reson Imaging 34:455–461
Coronado I, Gabr RE, Narayana PA (2021) Deep learning segmentation of gadolinium-enhancing lesions in multiple sclerosis. Mult Scler 27:519–527
Filippi M, van Waesberghe JH, Horsfield MA et al (1997) Interscanner variation in brain MRI lesion load measurements in MS: implications for clinical trials. Neurology 49:371–377
Commowick O, Istace A, Kain M et al (2018) Objective evaluation of multiple sclerosis lesion segmentation using a data management and processing infrastructure. Sci Rep 8:13650
Gabr RE, Lincoln JA, Kamali A et al (2020) Sensitive detection of infratentorial and upper cervical cord lesions in multiple sclerosis with combined 3D FLAIR and T2-weighted (FLAIR3) imaging. AJNR Am J Neuroradiol 41:2062–2067
Traboulsee A, Simon JH, Stone L et al (2016) Revised recommendations of the consortium of MS centers task force for a standardized MRI protocol and clinical guidelines for the diagnosis and follow-up of multiple sclerosis. AJNR Am J Neuroradiol 37:394–401
Funding
This work was partially funded by Zentrales Innovationsprogramm Mittelstand (ZIM) (contract number ZF4268403TS9) and by Hamburgische Investitions- und Förderbank (IFB) (contract number 51084589).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Roland Opfer.
Conflict of interest
Sven Schippling reports compensation for consulting, serving on scientific advisory boards, speaking, or other activities from Biogen, Celgene, Merck, Sanofi, and TEVA. Sven Schippling is currently an employee of Roche, Basel. Julia Krüger, Lothar Spies, and Roland Opfer are employees of jung diagnostics GmbH. Hagen H. Kitzler has received travel grants, speaker’s honoraria, financial research support, and consultancy fees from Bayer, Biogen Idec, Novartis, Siemens, and TEVA. He served on advisory boards for Biogen, Novartis, and Ixico. He received research grants from Novartis.
Statistics and biometry
One of the authors has a PhD in mathematics and a significant statistical expertise.
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was obtained from all subjects (patients) in this study.
Ethical approval
The need for written informed consent was waived by the ethics review board of the general medical council of the state of Hamburg, Germany (reference number 2021–300047-WF).
Study subjects or cohorts overlap
Part of the data used in our publication is publically available under https://www.sciencedirect.com/science/article/abs/pii/S1053811916303421 However, the compared data itself (manually/automatically segmented masks of lesions) were produced specifically for this paper. And therefore, no overlap to other studies is given.
Methodology
• retrospective.
• observational/experimental.
• multicenter study.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sven Schippling and Roland Opfer contributed equally as senior authors.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Krüger, J., Ostwaldt, AC., Spies, L. et al. Infratentorial lesions in multiple sclerosis patients: intra- and inter-rater variability in comparison to a fully automated segmentation using 3D convolutional neural networks. Eur Radiol 32, 2798–2809 (2022). https://doi.org/10.1007/s00330-021-08329-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-021-08329-3