Abstract
Objectives
The interpretability of convolutional neural networks (CNNs) for classifying subsolid nodules (SSNs) is insufficient for clinicians. Our purpose was to develop CNN models to classify SSNs on CT images and to investigate image features associated with the CNN classification.
Methods
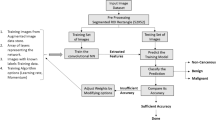
CT images containing SSNs with a diameter of ≤ 3 cm were retrospectively collected. We trained and validated CNNs by a 5-fold cross-validation method for classifying SSNs into three categories (benign and preinvasive lesions [PL], minimally invasive adenocarcinoma [MIA], and invasive adenocarcinoma [IA]) that were histologically confirmed or followed up for 6.4 years. The mechanism of CNNs on human-recognizable CT image features was investigated and visualized by gradient-weighted class activation map (Grad-CAM), separated activation channels and areas, and DeepDream algorithm.
Results
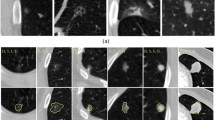
The accuracy was 93% for classifying 586 SSNs from 569 patients into three categories (346 benign and PL, 144 MIA, and 96 IA in 5-fold cross-validation). The Grad-CAM successfully located the entire region of image features that determined the final classification. Activated areas in the benign and PL group were primarily smooth margins (p < 0.001) and ground-glass components (p = 0.033), whereas in the IA group, the activated areas were mainly part-solid (p < 0.001) and solid components (p < 0.001), lobulated shapes (p < 0.001), and air bronchograms (p < 0.001). However, the activated areas for MIA were variable. The DeepDream algorithm showed the image features in a human-recognizable pattern that the CNN learned from a training dataset.
Conclusion
This study provides medical evidence to interpret the mechanism of CNNs that helps support the clinical application of artificial intelligence.
Key Points
• CNN achieved high accuracy (93%) in classifying subsolid nodules on CT images into three categories: benign and preinvasive lesions, MIA, and IA.
• The gradient-weighted class activation map (Grad-CAM) located the entire region of image features that determined the final classification, and the visualization of the separated activated areas was consistent with radiologists’ expertise for diagnosing subsolid nodules.
• DeepDream showed the image features that CNN learned from a training dataset in a human-recognizable pattern.







Similar content being viewed by others
Abbreviations
- AAH:
-
Atypical adenomatous hyperplasia
- AIS:
-
Adenocarcinoma in situ
- AUC:
-
Area under the ROC curve
- BMI:
-
Body mass index
- CNN:
-
Convolutional neural network
- CT:
-
Computed tomography
- Grad-CAM:
-
Gradient-weighted class activation map
- IA:
-
Invasive adenocarcinoma
- MIA:
-
Minimally invasive adenocarcinoma
- PL:
-
Preinvasive lesions
- ROC:
-
Receiver operating characteristic curve
- SSN:
-
Subsolid nodule
References
de Koning HJ, van der Aalst CM, de Jong PA et al (2020) Reduced lung-cancer mortality with volume CT screening in a randomized trial. N Engl J Med 382:503–513
Lee HW, Jin KN, Lee JK et al (2019) Long-term follow-up of ground-glass nodules after 5 years of stability. J Thorac Oncol 14:1370–1377
Silva M, Prokop M, Jacobs C et al (2018) Long-term active surveillance of screening detected subsolid nodules is a safe strategy to reduce overtreatment. J Thorac Oncol 13:1454–1463
McWilliams A, Tammemagi MC, Mayo JR et al (2013) Probability of cancer in pulmonary nodules detected on first screening CT. N Engl J Med 369:910–919
Henschke CI, Yip R, Yankelevitz DF et al (2013) Definition of a positive test result in computed tomography screening for lung cancer: a cohort study. Ann Intern Med 158:246–252
Travis WD, Brambilla E, Noguchi M et al (2011) International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 6:244–285
Naidich DP, Bankier AA, MacMahon H et al (2013) Recommendations for the management of subsolid pulmonary nodules detected at CT: a statement from the Fleischner Society. Radiology 266:304–317
Zhao W, Yang J, Sun Y et al (2018) 3D deep learning from CT scans predicts tumor invasiveness of subcentimeter pulmonary adenocarcinomas. Cancer Res 78:6881–6889
Wang S, Wang R, Zhang S et al (2018) 3D convolutional neural network for differentiating pre-invasive lesions from invasive adenocarcinomas appearing as ground-glass nodules with diameters ≤3 cm using HRCT. Quant Imaging Med Surg 8:491–499
Gong J, Liu J, Hao W et al (2020) A deep residual learning network for predicting lung adenocarcinoma manifesting as ground-glass nodule on CT images. Eur Radiol 30:1847–1855
Qin ZW, Yu FX, Liu CC, Chen X (2018) How convolutional neural networks see the world - a survey of convolutional neural network visualization methods. Mathematical Foundations of Computing 1:149–180
Zeiler MD, Fergus R (2014) Visualizing and understanding convolutional networks. Computer Vision - ECCV 2014 8689:818–833
Alexander M, Christopher O, Tyka M (2015) Inceptionism: Going deeper into neural networks. Available via http://ai.googleblog.com/2015/06/inceptionism-going-deeper-into-neural.html. Accessed 1 Feb 2021
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D (2016) Grad-CAM: Visual explanations from deep networks via gradient-based localization. Int J Comput Vis 2:336–359
Yamashita R, Nishio M, Do RKG, Togashi K (2018) Convolutional neural networks: an overview and application in radiology. Insights Imaging 9:611–629
Li JX, Xia TT, Yang XG et al (2018) Malignant solitary pulmonary nodules: assessment of mass growth rate and doubling time at follow-up CT. J Thorac Dis 10:S797–S806
Alpert JB, Ko JP (2018) Management of incidental lung nodules: Current strategy and rationale. Radiol Clin North Am 56:339–351
Xie X, Heuvelmans MA, van Ooijen PM, Oudkerk M, Vliegenthart R (2013) A practical approach to radiological evaluation of CT lung cancer screening examinations. Cancer Imaging 13:391–399
Horeweg N, Scholten ET, de Jong PA et al (2014) Detection of lung cancer through low-dose CT screening (NELSON): a prespecified analysis of screening test performance and interval cancers. Lancet Oncol 15:1342–1350
MacMahon H, Naidich DP, Goo JM et al (2017) Guidelines for management of incidental pulmonary nodules detected on ct images: from the Fleischner Society 2017. Radiology 284:228–243
Blagus R, Lusa L (2015) Joint use of over- and under-sampling techniques and cross-validation for the development and assessment of prediction models. BMC Bioinformatics 16:363
Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A (2016) Learning deep features for discriminative localization. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE 2921–2929
Nair A, Bartlett EC, Walsh SLF et al (2018) Variable radiological lung nodule evaluation leads to divergent management recommendations. Eur Respir J 52:1801359
Goutte C, Gaussier E (2005) A probabilistic interpretation of precision, recall and F-score, with implication for evaluationProceedings of the 27th European conference on Advances in Information Retrieval Research 345–359
Son JY, Lee HY, Kim JH et al (2016) Quantitative CT analysis of pulmonary ground-glass opacity nodules for distinguishing invasive adenocarcinoma from non-invasive or minimally invasive adenocarcinoma: the added value of using iodine mapping. Eur Radiol 26:43–54
Yanagawa M, Niioka H, Hata A et al (2019) Application of deep learning (3-dimensional convolutional neural network) for the prediction of pathological invasiveness in lung adenocarcinoma: a preliminary study. Medicine (Baltimore) 98:e16119
Kim H, Park CM, Koh JM, Lee SM, Goo JM (2014) Pulmonary subsolid nodules: what radiologists need to know about the imaging features and management strategy. Diagn Interv Radiol 20:47–57
Zhang Q, Cao R, Shi F, Nian Wu Y, Zhu S-C (2017) Interpreting CNN knowledge via an explanatory graph. arXiv e-prints. Available via https://arxiv.org/abs/1708.01785. Accessed 1 Feb 2021
Olah C, Mordvintsev A, Schubert L (2017) Feature visualization. Distill. https://doi.org/10.23915/distill.00007
DeepDreaming with TensorFlow (2016) Available via https://colab.research.google.com/github/tensorflow/examples/blob/master/community/en/r1/deepdream.ipynb. Accessed 1 Feb 2021
Olah C, Satyanarayan A, Johnson I et al (2018) The building blocks of interpretability. Distill. https://doi.org/10.23915/distill.00010
Schubert L, Petrov M, Carter S, Cammarata N, Goh G, Olah C (2020) OpenAI microscope. OpenAI. Available via https://openai.com/blog/microscope/. Accessed 1 Feb 2021
Coudray N, Ocampo PS, Sakellaropoulos T et al (2018) Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 24:1559–1567
Wang S, Shi J, Ye Z et al (2019) Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J 53:1800986
Nakkiran P, Kaplun G, Bansal Y, Yang T, Barak B, Sutskever I (2019) Deep double descent: where bigger models and more data hurt. arXiv e-prints. Available via https://arxiv.org/abs/1912.02292. Accessed 1 Feb 2021
Funding
This study has received funding from the National Natural Science Foundation of China (project no. 81971612), Ministry of Science and Technology of China (2016YFE0103000), Shanghai Municipal Education Commission – Gaofeng Clinical Medicine Grant Support (20181814), Shanghai Jiao Tong University (ZH2018ZDB10), and Clinical Research Innovation Plan of Shanghai General Hospital (CTCCR-2018B04, CTCCR-2019D05). The funders played no role in the study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Xueqian Xie.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained (No. SGH-2018-56).
Study subjects or cohorts overlap
No study subjects or cohorts have been previously reported.
Methodology
• Retrospective
• Diagnostic or prognostic study
• Performed at one institution
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 2539 kb)
Rights and permissions
About this article
Cite this article
Jiang, B., Zhang, Y., Zhang, L. et al. Human-recognizable CT image features of subsolid lung nodules associated with diagnosis and classification by convolutional neural networks. Eur Radiol 31, 7303–7315 (2021). https://doi.org/10.1007/s00330-021-07901-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-021-07901-1