Abstract
Objectives
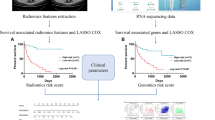
To develop a radiomics model using preoperative multiphasic CT for predicting distant metastasis after surgical resection in patients with localized clear cell renal cell carcinoma (ccRCC) and to identify key biological pathways underlying the predictive radiomics features using RNA sequencing data.
Methods
In this multi-institutional retrospective study, a CT radiomics metastasis score (RMS) was developed from a radiomics analysis cohort (n = 184) for distant metastasis prediction. Using a gene expression analysis cohort (n = 326), radiomics-associated gene modules were identified. Based on a radiogenomics discovery cohort (n = 42), key biological pathways were enriched from the gene modules. Furthermore, a multigene signature associated with RMS was constructed and validated on an independent radiogenomics validation cohort (n = 37).
Results
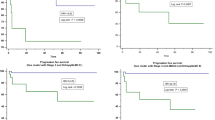
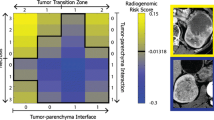
The 9-feature-based RMS predicted distant metastasis with an AUC of 0.861 in validation set and was independent with clinical factors (p < 0.001). A gene module comprising 114 genes was identified to be associated with all nine radiomics features (p < 0.05). Four enriched pathways were identified, including ECM-receptor interaction, focal adhesion, protein digestion and absorption, and PI3K-Akt pathways. Most of them play important roles in tumor progression and metastasis. A 19-gene signature was constructed from the radiomics-associated gene module and predicted metastasis with an AUC of 0.843 in the radiogenomics validation cohort.
Conclusions
CT radiomics features can predict distant metastasis after surgical resection of localized ccRCC while the predictive radiomics phenotypes may be driven by key biological pathways related to cancer progression and metastasis.
Key Points
• Radiomics features from primary tumor in preoperative CT predicted distant metastasis after surgical resection in patients with localized ccRCC.
• CT radiomics features predictive of distant metastasis were associated with key signaling pathways related to tumor progression and metastasis.
• Gene signature associated with radiomics metastasis score predicted distant metastasis in localized ccRCC.





Similar content being viewed by others
Abbreviations
- ccRCC:
-
Clear cell renal cell carcinoma
- CM:
-
Corticomedullary phase
- ECM:
-
Extracellular matrix
- FDR:
-
False discovery rate
- GLCM:
-
Gray level co-occurrence matrix
- GLDM:
-
Gray level dependency matrix
- GLRLM:
-
Gray level run length matrix
- GLSZM:
-
Gray level size matrix
- KEGG:
-
KOBAS-Kyoto Encyclopedia of Genes and Genomes
- LASSO:
-
Least absolute shrinkage and selection operator
- NG:
-
Nephrographic phase
- NGTDM:
-
Neighborhood gray tone difference matrix
- PI3K:
-
Phosphoinositide 3-kinases
- RMS:
-
Radiomics metastasis score
- Se:
-
Sensitivity
- Sp:
-
Specificity
- TCGA:
-
The cancer genome atlas
- TCIA:
-
The cancer imaging archive
- UE:
-
Unenhanced scans
- WGCNA:
-
Weighted gene co-expression network analysis
References
Capitanio U, Montorsi F (2016) Renal cancer. Lancet 387:894–906
Hsieh JJ, Purdue MP, Signoretti S et al (2017) Renal cell carcinoma. Nat Rev Dis Primers 3:17009
Ljungberg B, Bensalah K, Canfield S et al (2015) EAU guidelines on renal cell carcinoma: 2014 update. Eur Urol 67:913–924
Escudier B, Porta C, Schmidinger M et al (2016) Renal cell carcinoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 25:iii49–iii56
Lalani AKA, McGregor BA, Albiges L et al (2019) Systemic treatment of metastatic clear cell renal cell carcinoma in 2018: current paradigms, use of immunotherapy, and future directions. Eur Urol 75:100–110
Rydzanicz M, Wrzesinski T, Bluyssen HA et al (2013) Genomics and epigenomics of clear cell renal cell carcinoma: recent developments and potential applications. Cancer Lett 341:111–126
Randall JM, Millard F, Kurzrock R (2014) Molecular aberrations, targeted therapy, and renal cell carcinoma: current state-of-the-art. Cancer Metastasis Rev 33:1109–1124
Jones J, Out H, Spentzos D et al (2005) Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res 11:5730–5739
Jiang Z, Chu PG, Woda BA et al (2006) Analysis of RNA-binding protein IMP3 to predict metastasis and prognosis of renal-cell carcinoma: a retrospective study. Lancet Oncol 7:556–564
Lambin P, Leijenaar RTH, Deist TM et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14:749
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577
Huang YQ, Liang CH, He L et al (2016) Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol 34:2157–2164
Li J, Fang MJ, Wang R et al (2018) Diagnostic accuracy of dual-energy CT-based nomograms to predict lymph node metastasis in gastric cancer. Eur Radiol 28:5241–5249
Hu TD, Wang SP, Huang L et al (2019) A clinical radiomics nomogram for the preoperative prediction of lung metastasis in colorectal cancer patients with indeterminate pulmonary nodules. Eur Radiol 29:439–449
Coroller TP, Grossmann P, Hou Y et al (2015) CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol 114:345–350
Peeken JC, Bernhofer M, Spraker MB et al (2019) CT-based radiomic features predict tumor grading and have prognostic value in patients with soft tissue sarcomas treated with neoadjuvant radiation therapy. Radiother Oncol 135:187–196
Li ZC, Zhai GT, Zhang JH et al (2019) Differentiation of clear cell and non-clear cell renal cell carcinomas by all-relevant radiomics features from multiphase CT: a VHL mutation perspective. Eur Radiol 29:3996–4007
Timmeren JE, Leijenaar RTH, Elmpt W et al (2017) Survival prediction of non-small cell lung cancer patients using radiomics analyses of cone-beam CT images. Radiother Oncol 123:363–369
Antunes J, Viswanath S, Rusu M et al (2016) Radiomics analysis on FLT-PET/MRI for characterization of early treatment response in renal cell carcinoma: a proof-of-concept study. Transl Oncol 9:155–162
Beig N, Bera K, Prasanna P et al (2020) Radiogenomic-based survival risk stratification of tumor habitat on Gd-T1w MRI is associated with biological processes in glioblastoma. Clin Cancer Res 26:1866–1876
Wu J, Li B, Sun X et al (2017) Heterogeneous enhancement patterns of tumor-adjacent parenchyma at MR imaging are associated with dysregulated signaling pathways and poor survival in breast cancer. Radiology 285:401–413
Zhou M, Leung A, Echegaray S et al (2018) Non–small cell lung cancer radiogenomics map identifies relationships between molecular and imaging phenotypes with prognostic implications. Radiology 286:307–315
van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107
Kursa MB, Rudnicki WR (2010) Feature selection with the Boruta package. J Stat Softw 36:1–13
Tibshirani R (1997) The lasso method for variable selection in the cox model. Stat Med 16:385–395
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287
Karlo CA, Paolo PLD, Chaim J et al (2013) Radiogenomics of clear cell renal cell carcinoma: associations between CT imaging features and mutations. Radiology 270:464–471
Qian Z, Li Y, Sun Z et al (2018) Radiogenomics of lower-grade gliomas: a radiomic signature as a biological surrogate for survival prediction. Aging (Albany NY) 10:2884–2899
Gilkes DM, Semenza GL, Wirtz D (2014) Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer 14:430–439
Hynes RO (2009) Extracellular matrix: not just pretty fibrils. Science 326:1216–1219
Wirtz D, Konstantopoulos K, Searson PC (2011) The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat Rev Cancer 11:512–522
Mclean GW, Carragher NO, Avizienyte E, Evans J, Brunton VG, Frame MC (2005) The role of focal-adhesion kinase in cancer—a new therapeutic opportunity. Nat Rev Cancer 5:505–515
Scelo G, Riazalhosseini Y, Greger L et al (2014) Variation in genomic landscape of clear cell renal cell carcinoma across Europe. Nat Commun 5:5135
Guo H, German P, Bai S et al (2015) The PI3K/AKT pathway and renal cell carcinoma. J Genet Genomics 42:343–353
Schultz L, Chaux A, Albadine R et al (2011) Immunoexpression status and prognostic value of mTOR and hypoxia-induced pathway members in primary and metastatic clear cell renal cell carcinomas. Am J Surg Pathol 35:1549–1556
Akkiprik M, Peker I, Özmen T et al (2015) A: Identification of differentially expressed IGFBP5-related genes in breast cancer tumor tissues using cDNA microarray experiments. Genes (Basel) 6:1201–1214
Wang DD, Liu ZW, Han MM et al (2015) Microarray based analysis of gene expression patterns in pancreatic neuroendocrine tumors. Eur Rev Med Pharmacol 19:3367–3374
Funding
This study has received funding by National Natural Science Foundation of China (61901458), China Postdoctoral Science Foundation (2019 M663187), Youth Innovation Promotion Association of Chinese Academy of Sciences (2018364), and Shenzhen Basic Research Program (JCYJ20170413162354654).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The guarantor of this publication is Zhi-Cheng Li.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors (Zhi-Cheng Li) has significant statistical expertise.
Informed consent
Written informed consent was waved by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Study subjects or cohorts overlap
None.
Methodology
• retrospective
• diagnostic or prognostic study
• performed at multiple institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
ESM 1
(DOCX 139 kb)
Rights and permissions
About this article
Cite this article
Zhao, Y., Liu, G., Sun, Q. et al. Validation of CT radiomics for prediction of distant metastasis after surgical resection in patients with clear cell renal cell carcinoma: exploring the underlying signaling pathways. Eur Radiol 31, 5032–5040 (2021). https://doi.org/10.1007/s00330-020-07590-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07590-2