Abstract
Objectives
To evaluate the prediction performance of deep convolutional neural network (DCNN) based on ultrasound (US) images for the assessment of breast cancer molecular subtypes.
Methods
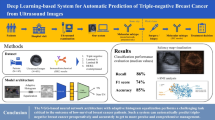
A dataset of 4828 US images from 1275 patients with primary breast cancer were used as the training samples. DCNN models were constructed primarily to predict the four St. Gallen molecular subtypes and secondarily to identify luminal disease from non-luminal disease based on the ground truth from immunohistochemical of whole tumor surgical specimen. US images from two other institutions were retained as independent test sets to validate the system. The models’ performance was analyzed using per-class accuracy, positive predictive value (PPV), and Matthews correlation coefficient (MCC).
Results
The model achieved good performance in identifying the four breast cancer molecular subtypes in the two test sets, with accuracy ranging from 80.07% (95% CI, 76.49–83.23%) to 97.02% (95% CI, 95.22–98.16%) and 87.94% (95% CI, 85.08–90.31%) to 98.83% (95% CI, 97.60–99.43) for the two test cohorts for each sub-category, respectively. In terms of 4-class weighted average MCC, the model achieved 0.59 for test cohort A and 0.79 for test cohort B. Specifically, the DCNN also yielded good diagnostic performance in discriminating luminal disease from non-luminal disease, with a PPV of 93.29% (95% CI, 90.63–95.23%) and 88.21% (95% CI, 85.12–90.73%) for the two test cohorts, respectively.
Conclusion
Using pretreatment US images of the breast cancer, deep learning model enables the assessment of molecular subtypes with high diagnostic accuracy.
Trial registration
Clinical trial number: ChiCTR1900027676
Key Points
• Deep convolutional neural network (DCNN) helps clinicians assess tumor features with accuracy.
• Multicenter retrospective study shows that DCNN derived from pretreatment ultrasound imagine improves the prediction of breast cancer molecular subtypes.
• Management of patients becomes more precise based on the DCNN model.





Similar content being viewed by others
Abbreviations
- AUC:
-
Area under the receiver operator characteristic curve
- CI:
-
Confidence interval
- DCNN:
-
Deep convolutional neural network
- ER:
-
Estrogen receptor
- HER2:
-
Human epidermal growth factor receptor 2
- IHC:
-
Immunohistochemical
- MCC:
-
Matthews correlation coefficient
- PR:
-
Progesterone receptor
- ROC:
-
Receiver operating characteristic curve
References
Chen W, Zheng R, Baade PD et al (2016) Cancer statistics in China, 2015. CA Cancer J Clin 66:115–132
Haynes B, Sarma A, Nangia-Makker P, Shekhar MP (2017) Breast cancer complexity: implications of intratumoral heterogeneity in clinical management. Cancer Metastasis Rev 36:547–555
Zardavas D, Irrthum A, Swanton C, Piccart M (2015) Clinical management of breast cancer heterogeneity. Nat Rev Clin Oncol 12:381–394
Martelotto LG, Ng CK, Piscuoglio S, Weigelt B, Reis-Filho JS (2014) Breast cancer intra-tumor heterogeneity. Breast Cancer Res 16:210
Sorlie T, Perou CM, Tibshirani R et al (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 98:10869–10874
Prat A, Pineda E, Adamo B et al (2015) Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast 24(Suppl 2):S26–S35
Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature 490:61–70
Prat A, Cheang MCU, Martín M et al (2013) Prognostic significance of progesterone receptor–positive tumor cells within immunohistochemically defined luminal A breast cancer. J Clin Oncol 31:203–209
Tsoutsou PG, Vozenin MC, Durham AD, Bourhis J (2017) How could breast cancer molecular features contribute to locoregional treatment decision making? Crit Rev Oncol Hematol 110:43–48
Ahn HJ, Jung SJ, Kim TH, Oh MK, Yoon H (2015) Differences in clinical outcomes between luminal A and B type breast cancers according to the St. Gallen consensus 2013. J Breast Cancer 18:149–159
Spratt DE, Evans MJ, Davis BJ et al (2015) Androgen receptor upregulation mediates radioresistance after ionizing radiation. Cancer Res 75:4688–4696
Li X, Zhang S, Zhang Q et al (2019) Diagnosis of thyroid cancer using deep convolutional neural network models applied to sonographic images: a retrospective, multicohort, diagnostic study. Lancet Oncol 20:193–201
Ehteshami Bejnordi B, Veta M, Johannes Van Diest P et al (2017) Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318:2199–2210
Zhou LQ, Wu XL, Huang SY et al (2020) Lymph node metastasis prediction from primary breast cancer US images using deep learning. Radiology 294:19–28
Fujioka T, Mori M, Kubota K et al (2019) Breast ultrasound image synthesis using deep convolutional generative adversarial networks. Diagnostics (Basel) 9:176
Xiao T, Liu L, Li K, Qin W, Yu S, Li Z (2018) Comparison of transferred deep neural networks in ultrasonic breast masses discrimination. Biomed Res Int 2018:4605191–4605199
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444
Goldhirsch A, Winer EP, Coates AS et al (2013) Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol 24:2206–2223
Curigliano G, Burstein HJ, Winer EP et al (2017) De-escalating and escalating treatments for early-stage breast cancer: the St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann Oncol 28:1700–1712
He K, Gkioxari G, Dollar P, Girshick R (2018) Mask R-CNN. IEEE Trans Pattern Anal Mach Intell 2018:1
Redmon J, Farhadi A. YOLO9000: better, faster, stronger. Proc IEEE Conf Comput Vis Pattern Recognit 2017; published online Nov 9. https://doi.org/10.1109/CVPR.2017.690
Xie Y, Xia Y, Zhang J et al (2019) Knowledge-based collaborative deep learning for benign-malignant lung nodule classification on chest CT. IEEE Trans Med Imaging 38:991–1004
Rajpurkar P, Irvin J, Ball RL et al (2018) Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med 15:e1002686
Lin T, Goyal P, Girshick R, He K, Dollar P (2020) Focal loss for dense object detection. IEEE Trans Pattern Anal Mach Intell 42:318–327
Esteva A, Kuprel B, Novoa RA et al (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542:115–118
Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A (2016) Learning deep features for discriminative localization. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 2921–2929
Hand DJ, Till RJ (2001) A simple generalisation of the area under the ROC curve for multiple class classification problems. Mach Learn 45:171–186
Hannun AY, Rajpurkar P, Haghpanahi M et al (2019) Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med 25:65–69
Zhao W, Yang J, Sun Y et al (2018) 3D deep learning from CT scans predicts tumor invasiveness of subcentimeter pulmonary adenocarcinomas. Cancer Res 78:6881–6889
Waks AG, Winer EP (2019) Breast cancer treatment: a review. JAMA 321:288–300
Wiechmann L, Sampson M, Stempel M et al (2009) Presenting features of breast cancer differ by molecular subtype. Ann Surg Oncol 16:2705–2710
Smid M, Wang Y, Zhang Y et al (2008) Subtypes of breast cancer show preferential site of relapse. Cancer Res 68:3108–3114
Chen XS, Wu JY, Huang O et al (2010) Molecular subtype can predict the response and outcome of Chinese locally advanced breast cancer patients treated with preoperative therapy. Oncol Rep 23:1213–1220
Kyndi M, Sørensen FB, Knudsen H, Overgaard M, Nielsen HM, Overgaard J (2008) Estrogen receptor, progesterone receptor, HER-2, and response to postmastectomy radiotherapy in high-risk breast cancer: the Danish Breast Cancer Cooperative Group. J Clin Oncol 26:1419–1426
Tran B, Bedard PL (2011) Luminal-B breast cancer and novel therapeutic targets. Breast Cancer Res 13:221
Huber KE, Carey LA, Wazer DE (2009) Breast cancer molecular subtypes in patients with locally advanced disease: impact on prognosis, patterns of recurrence, and response to therapy. Semin Radiat Oncol 19:204–210
Liu H, Wan J, Xu G et al (2019) Conventional US and 2-D shear wave elastography of virtual touch tissue imaging quantification: correlation with immunohistochemical subtypes of breast cancer. Ultrasound Med Biol 45:2612–2622
Rashmi S, Kamala S, Murthy SS, Kotha S, Rao YS, Chaudhary KV (2018) Predicting the molecular subtype of breast cancer based on mammography and ultrasound findings. Indian J Radiol Imaging 28:354–361
Mazurowski MA, Zhang J, Grimm LJ, Yoon SC, Silber JI (2014) Radiogenomic analysis of breast cancer: luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology 273:365–372
Presta M, Dell Era P, Mitola S, Moroni E, Ronca R, Rusnati M (2005) Fibroblast growth factor/fibroblast growth factor receptor system in angiogenesis. Cytokine Growth Factor Rev 16:159–178
Casanovas O, Hicklin DJ, Bergers G, Hanahan D (2005) Drug resistance by evasion of antiangiogenic targeting of VEGF signaling in late-stage pancreatic islet tumors. Cancer Cell 8:299–309
Zhu Z, Albadawy E, Saha A, Zhang J, Harowicz MR, Mazurowski MA (2019) Deep learning for identifying radiogenomic associations in breast cancer. Comput Biol Med 109:85–90
Cejalvo JM, Pascual T, Fernández-Martínez A et al (2018) Clinical implications of the non-luminal intrinsic subtypes in hormone receptor-positive breast cancer. Cancer Treat Rev 67:63–70
Acknowledgements
The authors thank all radiologists of the three hospitals for assisting with collection of the imaging data used in this study.
Funding
This study has received funding by grant from the Project funded by China Postdoctoral Science Foundation (2020M682422), National Natural Science Foundation of China (No. 61976012), Wuhan Science and Technology Bureau (No. 2017060201010181), Health Commission of Hubei Province (WJ2019M077, WJ2019H227), Shihezi Science and Technology Bureau (2019ZH11), Key project supported by the Xinjiang Construction Corps (2019DB012), and Natural Science Foundation of Hubei Province (2019CFB286).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Professor Xin-Wu Cui.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Only if the study is on human subjects:
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• multicenter study
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 430 kb)
Rights and permissions
About this article
Cite this article
Jiang, M., Zhang, D., Tang, SC. et al. Deep learning with convolutional neural network in the assessment of breast cancer molecular subtypes based on US images: a multicenter retrospective study. Eur Radiol 31, 3673–3682 (2021). https://doi.org/10.1007/s00330-020-07544-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07544-8