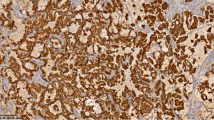
Abstract
Objectives
To develop a nomogram to identify anaplastic lymphoma kinase (ALK) mutations in lung adenocarcinoma patients using clinical, CT, PET/CT, and histopathological features.
Methods
This retrospective study included 399 lung adenocarcinoma patients (129 ALK-rearranged patients and 270 ALK-negative patients) that were randomly divided into a training cohort and an internal validation cohort (4:1 ratio). Clinical factors, radiologist-defined CT features, maximum standard uptake values (SUVmax), and histopathological features were used to construct predictive models with stepwise backward-selection multivariate logistic regression (MLR). The models were then evaluated using the AUC. The integrated model was compared to the clinico-radiological model using the DeLong test to evaluate the role of histopathological features. An associated individualized nomogram was established.
Results
The integrated model reached an AUC of 0.918 (95% CI, 0.886–0.950), sensitivity of 0.774, and specificity of 0.934 in the training cohort and an AUC of 0.857 (95% CI, 0.777–0.937), sensitivity of 0.739, and specificity of 0.810 in the validation cohort. The MLR analysis showed that younger age, never smoker, lymph node enlargement, the presence of cavity, high SUVmax, solid or micropapillary predominant histology subtype, and local invasiveness were strong and independent predictors of ALK rearrangements. The nomogram calculated the risk of harboring ALK mutation for lung adenocarcinoma patients and exhibited a good generalization ability.
Conclusion
Our study demonstrates that histopathological features added value to the imaging characteristics-based model. The nomogram with clinical, imaging, and histopathological features can serve as a supplementary non-invasive tool to evaluate the probability of ALK rearrangement in lung adenocarcinoma.
Key Points
• The developed nomogram can accurately predict the probability of lung adenocarcinoma harboring ALK-fused gene.
• Pathological analysis is important to predict ALK rearrangement in lung adenocarcinoma.
• Lung adenocarcinoma with lepidic predominant growth pattern and TTF-1 negativity is unlikely to have ALK rearrangement.






Similar content being viewed by others
Abbreviations
- AIS:
-
Adenocarcinoma in situ
- APA:
-
Acinar predominant adenocarcinoma
- AUC:
-
Area under the curve
- CI:
-
Confidence interval
- CT:
-
Computed tomography
- DICOM:
-
Digital imaging and communications in medicine
- EML4:
-
Echinoderm microtubule associated protein-like 4
- GGO:
-
Ground-glass opacity
- IAC:
-
Invasive adenocarcinoma
- IMA:
-
Invasive mucinous adenocarcinoma
- LPA:
-
Lepidic predominant adenocarcinoma
- MIA:
-
Minimally invasive adenocarcinoma
- MLR:
-
Multivariate logistic regression
- MPA:
-
Micropapillary predominant adenocarcinoma
- NCCN:
-
National Comprehensive Cancer Network
- NSCLC:
-
Non-small-cell lung cancer
- PPA:
-
Papillary predominant adenocarcinoma
- ROC:
-
Receiver operating characteristic
- SPA:
-
Solid predominant adenocarcinoma
References
Miranda-Filho A, Pineros M, Bray F (2019) The descriptive epidemiology of lung cancer and tobacco control: a global overview 2018. Salud Publica Mex 61:219–229
Duma N, Santana-Davila R, Molina JR (2019) Non-small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc 94:1623–1640
National Comprehensive Cancer Network (NCCN) Non-small cell lung cancer (Version 3. 2020). Available via https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf. Accessed 2 Mar 2020
Zito Marino F, Liguori G, Aquino G et al (2015) Intratumor heterogeneity of ALK-rearrangements and homogeneity of EGFR-mutations in mixed lung adenocarcinoma. PLoS One. https://doi.org/10.1371/journal.pone.0139264
Cai W, Lin D, Wu C et al (2015) Intratumoral heterogeneity of ALK-rearranged and ALK/EGFR coaltered lung adenocarcinoma. J Clin Oncol 33:3701–3709
de Sousa VML, Carvalho L (2018) Heterogeneity in lung cancer. Pathobiology 85:96–107
Jankovic R, Goncalves HJ, Cavic M et al (2019) LungCARD - report on worldwide research and clinical practices related to lung cancer. J BUON 24:11–19
Paolini D, Tiseo M, Demma F et al (2018) Ventana ALK (D5F3) in the detection of patients affected by anaplastic lymphoma kinase-positive non-small-cell lung cancer: clinical and budget effect. Clin Lung Cancer 19:e735–e743
Mendoza DP, Stowell J, Muzikansky A, Shepard J-AO, Shaw AT, Digumarthy SR (2019) Computed tomography imaging characteristics of non-small-cell lung cancer with anaplastic lymphoma kinase rearrangements: a systematic review and meta-analysis. Clin Lung Cancer 213:1059–1072
Jeong C, Lee H, Han J et al (2015) Role of imaging biomarkers in predicting anaplastic lymphoma kinase–positive lung adenocarcinoma. Clin Nucl Med 40:34–39
Yamamoto S, Korn RL, Oklu R et al (2014) ALK molecular phenotype in non-small cell lung cancer: CT radiogenomic characterization. Radiology 272:568–576
Zhao F, Xu M, Lei H et al (2015) Clinicopathological characteristics of patients with non-small-cell lung cancer who harbor EML4-ALK fusion gene: a meta-analysis. PLoS One. https://doi.org/10.1371/journal.pone.0117333
Rizzo S, Petrella F, Buscarino V et al (2016) CT radiogenomic characterization of EGFR, K-RAS, and ALK mutations in non-small cell lung cancer. Eur Radiol 26:32–42
Dong YJ, Cai YR, Zhou LJ et al (2016) Association between the histological subtype of lung adenocarcinoma, EGFR/KRAS mutation status and the ALK rearrangement according to the novel IASLC/ATS/ERS classification. Oncol Lett 11:2552–2558
Yoshida A, Tsuta K, Nakamura H et al (2011) Comprehensive histologic analysis of ALK-rearranged lung carcinomas. Am J Surg Pathol 35:1226–1234
Nishino M, Klepeis VE, Yeap BY et al (2012) Histologic and cytomorphologic features of ALK-rearranged lung adenocarcinomas. Mod Pathol 25:1462–1472
Amin M, Edge S, Greene F et al (2017) AJCC Cancer Staging Manual, 8th edn. Springer, New York
Travis W, Brambilla E, Noguchi M et al (2011) International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Classification of Lung Adenocarcinoma. J Thorac Oncol 6:244–285
Lindeman NI, Cagle PT, Aisner DL et al (2018) Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med 142:321–346
van Buuren S, Groothuis-Oudshoorn K (2011) mice: multivariate imputation by chained equations in R. Available via https://www.jstatsoft.org/article/view/v045i03. Accessed 2 Mar 2020
Donders AR, van der Heijden GJ, Stijnen T, Moons KG (2006) Review: a gentle introduction to imputation of missing values. J Clin Epidemiol 59:1087–1091
Heymans MW, van Buuren S, Knol DL, van Mechelen W, de Vet HC (2007) Variable selection under multiple imputation using the bootstrap in a prognostic study. BMC Med Res Methodol. https://doi.org/10.1186/1471-2288-7-33
Jokoji R, Yamasaki T, Minami S et al (2010) Combination of morphological feature analysis and immunohistochemistry is useful for screening of EML4-ALK-positive lung adenocarcinoma. J Clin Pathol 63:1066–1070
Choi C, Kim M, Hwang H, Lee J, Kim W (2015) Advanced adenocarcinoma of the lung: comparison of CT characteristics of patients with anaplastic lymphoma kinase gene rearrangement and those with epidermal growth factor receptor mutation. Radiology 275:272–279
Park J, Yamaura H, Yatabe Y et al (2014) Anaplastic lymphoma kinase gene rearrangements in patients with advanced-stage non-small-cell lung cancer: CT characteristics and response to chemotherapy. Cancer Med 3:118–123
Halpenny DF, Riely GJ, Hayes S et al (2014) Are there imaging characteristics associated with lung adenocarcinomas harboring ALK rearrangements? Lung Cancer 86:190–194
Seto K, Kuroda H, Yoshida T et al (2018) Higher frequency of occult lymph node metastasis in clinical N0 pulmonary adenocarcinoma with ALK rearrangement. Cancer Manag Res 10:2117–2124
Inamura K, Takeuchi K, Togashi Y et al (2009) EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod Pathol 22:508–515
Zhou JY, Zheng J, Yu ZF et al (2015) Comparative analysis of clinicoradiologic characteristics of lung adenocarcinomas with ALK rearrangements or EGFR mutations. Eur Radiol 25:1257–1266
Wang H, Schabath MB, Liu Y et al (2016) Clinical and CT characteristics of surgically resected lung adenocarcinomas harboring ALK rearrangements or EGFR mutations. Eur J Radiol 85:1934–1940
Kim TJ, Lee CT, Jheon SH, Park JS, Chung JH (2016) Radiologic characteristics of surgically resected non-small cell lung cancer with ALK rearrangement or EGFR mutations. Ann Thorac Surg 101:473–480
Yoon HJ, Sohn I, Cho JH et al (2015) Decoding tumor phenotypes for ALK, ROS1, and RET fusions in lung adenocarcinoma using a radiomics approach. Medicine (Baltimore). https://doi.org/10.1097/MD.0000000000001753
Song L, Zhu Z, Mao L et al (2020) Clinical, conventional CT and radiomic feature-based machine learning models for predicting ALK rearrangement status in lung adenocarcinoma patients. Front Oncol. https://doi.org/10.3389/fonc.2020.00369
Rolfo C, Mack PC, Scagliotti GV et al (2018) Liquid biopsy for advanced non-small cell lung cancer (NSCLC): a statement paper from the IASLC. J Thorac Oncol 13:1248–1268
Funding
This study has received funding from the Beijing Science and Technology Planning Project (Z201100005620008) and the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2019PT320008 and 2018PT32003).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Zheng-yu Jin.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
Dr. Wei Han (one of the authors) kindly provided statistical advice for this manuscript.
Informed consent
Written informed consent was waived by the Institutional Review Board of our institution due to the retrospective nature of the study.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• Retrospective
• Diagnostic or prognostic study
• Performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 622 kb)
Rights and permissions
About this article
Cite this article
Song, L., Zhu, Z., Wu, H. et al. Individualized nomogram for predicting ALK rearrangement status in lung adenocarcinoma patients. Eur Radiol 31, 2034–2047 (2021). https://doi.org/10.1007/s00330-020-07331-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07331-5