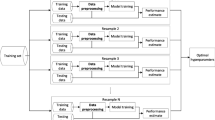
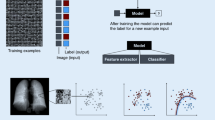
Abstract
In recent years, there has been a dramatic increase in research papers about machine learning (ML) and artificial intelligence in radiology. With so many papers around, it is of paramount importance to make a proper scientific quality assessment as to their validity, reliability, effectiveness, and clinical applicability. Due to methodological complexity, the papers on ML in radiology are often hard to evaluate, requiring a good understanding of key methodological issues. In this review, we aimed to guide the radiology community about key methodological aspects of ML to improve their academic reading and peer-review experience. Key aspects of ML pipeline were presented within four broad categories: study design, data handling, modelling, and reporting. Sixteen key methodological items and related common pitfalls were reviewed with a fresh perspective: database size, robustness of reference standard, information leakage, feature scaling, reliability of features, high dimensionality, perturbations in feature selection, class balance, bias-variance trade-off, hyperparameter tuning, performance metrics, generalisability, clinical utility, comparison with traditional tools, data sharing, and transparent reporting.
Key Points
• Machine learning is new and rather complex for the radiology community.
• Validity, reliability, effectiveness, and clinical applicability of studies on machine learning can be evaluated with a proper understanding of key methodological concepts about study design, data handling, modelling, and reporting.
• Understanding key methodological concepts will provide a better academic reading and peer-review experience for the radiology community.






Similar content being viewed by others
Abbreviations
- ML:
-
Machine learning
References
Choy G, Khalilzadeh O, Michalski M et al (2018) Current applications and future impact of machine learning in radiology. Radiology 288:318–328. https://doi.org/10.1148/radiol.2018171820
Wang S, Summers RM (2012) Machine learning and radiology. Med Image Anal 16:933–951. https://doi.org/10.1016/j.media.2012.02.005
Jordan MI, Mitchell TM (2015) Machine learning: trends, perspectives, and prospects. Science 349:255–260. https://doi.org/10.1126/science.aaa8415
Kohli M, Prevedello LM, Filice RW, Geis JR (2017) Implementing machine learning in radiology practice and research. AJR Am J Roentgenol 208:754–760. https://doi.org/10.2214/AJR.16.17224
Sollini M, Antunovic L, Chiti A, Kirienko M (2019) Towards clinical application of image mining: a systematic review on artificial intelligence and radiomics. Eur J Nucl Med Mol Imaging 46:2656–2672. https://doi.org/10.1007/s00259-019-04372-x
Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL (2018) Artificial intelligence in radiology. Nat Rev Cancer 18:500–510. https://doi.org/10.1038/s41568-018-0016-5
Do HM, Spear LG, Nikpanah M et al (2020) Augmented radiologist workflow improves report value and saves time: a potential model for implementation of artificial intelligence. Acad Radiol 27:96–105. https://doi.org/10.1016/j.acra.2019.09.014
Lou R, Lalevic D, Chambers C, Zafar HM, Cook TS (2020) Automated detection of radiology reports that require follow-up imaging using natural language processing feature engineering and machine learning classification. J Digit Imaging 33:131–136. https://doi.org/10.1007/s10278-019-00271-7
Mokrane F-Z, Lu L, Vavasseur A et al (2020) Radiomics machine-learning signature for diagnosis of hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules. Eur Radiol 30:558–570. https://doi.org/10.1007/s00330-019-06347-w
Schaffter T, Buist DSM, Lee CI et al (2020) Evaluation of combined artificial intelligence and radiologist assessment to interpret screening mammograms. JAMA Netw Open 3:e200265. https://doi.org/10.1001/jamanetworkopen.2020.0265
Chauvie S, De Maggi A, Baralis I et al (2020) Artificial intelligence and radiomics enhance the positive predictive value of digital chest tomosynthesis for lung cancer detection within SOS clinical trial. Eur Radiol. https://doi.org/10.1007/s00330-020-06783-z
Fischer AM, Varga-Szemes A, Martin SS et al (2020) Artificial intelligence-based fully automated per lobe segmentation and emphysema-quantification based on chest computed tomography compared with global initiative for chronic obstructive lung disease severity of smokers. J Thorac Imaging. https://doi.org/10.1097/RTI.0000000000000500
Kocak B, Durmaz ES, Ates E, Kaya OK, Kilickesmez O (2019) Unenhanced CT texture analysis of clear cell renal cell carcinomas: a machine learning-based study for predicting histopathologic nuclear grade. AJR Am J Roentgenol:W1–W8. https://doi.org/10.2214/AJR.18.20742
Kocak B, Durmaz ES, Ates E, Ulusan MB (2019) Radiogenomics in clear cell renal cell carcinoma: machine learning-based high-dimensional quantitative CT texture analysis in predicting PBRM1 mutation status. AJR Am J Roentgenol 212:W55–W63. https://doi.org/10.2214/AJR.18.20443
Kocak B, Durmaz ES, Ates E et al (2020) Radiogenomics of lower-grade gliomas: machine learning-based MRI texture analysis for predicting 1p/19q codeletion status. Eur Radiol 30:877–886. https://doi.org/10.1007/s00330-019-06492-2
Greffier J, Hamard A, Pereira F et al (2020) Image quality and dose reduction opportunity of deep learning image reconstruction algorithm for CT: a phantom study. Eur Radiol. https://doi.org/10.1007/s00330-020-06724-w
Parmar C, Barry JD, Hosny A, Quackenbush J, Aerts HJWL (2018) Data analysis strategies in medical imaging. Clin Cancer Res 24:3492–3499. https://doi.org/10.1158/1078-0432.CCR-18-0385
Thrall JH, Li X, Li Q et al (2018) Artificial intelligence and machine learning in radiology: opportunities, challenges, pitfalls, and criteria for success. J Am Coll Radiol 15:504–508. https://doi.org/10.1016/j.jacr.2017.12.026
Leek JT, Scharpf RB, Bravo HC et al (2010) Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet 11:733–739. https://doi.org/10.1038/nrg2825
Johnson WE, Li C, Rabinovic A (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8:118–127. https://doi.org/10.1093/biostatistics/kxj037
Quackenbush J (2002) Microarray data normalization and transformation. Nat Genet 32(Suppl):496–501. https://doi.org/10.1038/ng1032
Lee ML, Kuo FC, Whitmore GA, Sklar J (2000) Importance of replication in microarray gene expression studies: statistical methods and evidence from repetitive cDNA hybridizations. Proc Natl Acad Sci U S A 97:9834–9839. https://doi.org/10.1073/pnas.97.18.9834
Yu K-H, Beam AL, Kohane IS (2018) Artificial intelligence in healthcare. Nat Biomed Eng 2:719–731. https://doi.org/10.1038/s41551-018-0305-z
Koçak B, Durmaz EŞ, Ateş E, Kılıçkesmez Ö (2019) Radiomics with artificial intelligence: a practical guide for beginners. Diagn Interv Radiol 25:485–495. https://doi.org/10.5152/dir.2019.19321
Hernández B, Parnell A, Pennington SR (2014) Why have so few proteomic biomarkers “survived” validation? (sample size and independent validation considerations). Proteomics 14:1587–1592. https://doi.org/10.1002/pmic.201300377
Way TW, Sahiner B, Hadjiiski LM, Chan H-P (2010) Effect of finite sample size on feature selection and classification: a simulation study. Med Phys 37:907–920. https://doi.org/10.1118/1.3284974
Chan HP, Sahiner B, Wagner RF, Petrick N (1999) Classifier design for computer-aided diagnosis: effects of finite sample size on the mean performance of classical and neural network classifiers. Med Phys 26:2654–2668. https://doi.org/10.1118/1.598805
Sollini M, Cozzi L, Antunovic L, Chiti A, Kirienko M (2017) PET Radiomics in NSCLC: state of the art and a proposal for harmonization of methodology. Sci Rep 7:358. https://doi.org/10.1038/s41598-017-00426-y
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577. https://doi.org/10.1148/radiol.2015151169
Perlich C (2010) Learning curves in machine learning. In: Sammut C, Webb GI (eds) Encyclopedia of machine learning. Springer US, Boston, MA, pp 577–580
Krause J, Gulshan V, Rahimy E et al (2018) Grader variability and the importance of reference standards for evaluating machine learning models for diabetic retinopathy. Ophthalmology 125:1264–1272. https://doi.org/10.1016/j.ophtha.2018.01.034
Zwanenburg A (2019) Radiomics in nuclear medicine: robustness, reproducibility, standardization, and how to avoid data analysis traps and replication crisis. Eur J Nucl Med Mol Imaging 46:2638–2655. https://doi.org/10.1007/s00259-019-04391-8
Mwangi B, Tian TS, Soares JC (2014) A review of feature reduction techniques in neuroimaging. Neuroinformatics 12:229–244. https://doi.org/10.1007/s12021-013-9204-3
Zwanenburg A, Löck S (2018) Why validation of prognostic models matters? Radiother Oncol 127:370–373. https://doi.org/10.1016/j.radonc.2018.03.004
Huber W, von Heydebreck A, Sültmann H, Poustka A, Vingron M (2002) Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18(Suppl 1):S96–S104. https://doi.org/10.1093/bioinformatics/18.suppl_1.s96
Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep network training by reducing internal covariate shift. ArXiv150203167 Cs
Ba JL, Kiros JR, Hinton GE (2016) Layer normalization. ArXiv160706450 Cs stat
Kocak B, Ates E, Durmaz ES, Ulusan MB, Kilickesmez O (2019) Influence of segmentation margin on machine learning-based high-dimensional quantitative CT texture analysis: a reproducibility study on renal clear cell carcinomas. Eur Radiol 29:4765–4775. https://doi.org/10.1007/s00330-019-6003-8
Kocak B, Durmaz ES, Kaya OK, Ates E, Kilickesmez O (2019) Reliability of single-slice-based 2D CT texture analysis of renal masses: influence of intra- and interobserver manual segmentation variability on radiomic feature reproducibility. AJR Am J Roentgenol 213:377–383. https://doi.org/10.2214/AJR.19.21212
Koçak B (2019) Reliability of 2D magnetic resonance imaging texture analysis in cerebral gliomas: influence of slice selection bias on reproducibility of radiomic features. Istanb Med J 20:413–417
Um H, Tixier F, Bermudez D, Deasy JO, Young RJ, Veeraraghavan H (2019) Impact of image preprocessing on the scanner dependence of multi-parametric MRI radiomic features and covariate shift in multi-institutional glioblastoma datasets. Phys Med Biol 64:165011. https://doi.org/10.1088/1361-6560/ab2f44
Berenguer R, Pastor-Juan MDR, Canales-Vázquez J et al (2018) Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology 288:407–415. https://doi.org/10.1148/radiol.2018172361
Zhovannik I, Bussink J, Traverso A et al (2019) Learning from scanners: bias reduction and feature correction in radiomics. Clin Transl Radiat Oncol 19:33–38. https://doi.org/10.1016/j.ctro.2019.07.003
Bologna M, Corino V, Mainardi L (2019) Technical note: virtual phantom analyses for preprocessing evaluation and detection of a robust feature set for MRI-radiomics of the brain. Med Phys 46:5116–5123. https://doi.org/10.1002/mp.13834
He H, Garcia EA (2009) Learning from imbalanced data. IEEE Trans Knowl Data Eng 21:1263–1284
Meyer M, Ronald J, Vernuccio F et al (2019) Reproducibility of CT radiomic features within the same patient: influence of radiation dose and CT reconstruction settings. Radiology 293:583–591. https://doi.org/10.1148/radiol.2019190928
Qiu Q, Duan J, Duan Z et al (2019) Reproducibility and non-redundancy of radiomic features extracted from arterial phase CT scans in hepatocellular carcinoma patients: impact of tumor segmentation variability. Quant Imaging Med Surg 9:453–464. https://doi.org/10.21037/qims.2019.03.02
Owens CA, Peterson CB, Tang C et al (2018) Lung tumor segmentation methods: impact on the uncertainty of radiomics features for non-small cell lung cancer. PLoS One 13:e0205003. https://doi.org/10.1371/journal.pone.0205003
Estrada S, Lu R, Conjeti S et al (2020) FatSegNet: a fully automated deep learning pipeline for adipose tissue segmentation on abdominal Dixon MRI. Magn Reson Med 83:1471–1483. https://doi.org/10.1002/mrm.28022
Lambin P, Leijenaar RTH, Deist TM et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14:749–762. https://doi.org/10.1038/nrclinonc.2017.141
Leger S, Zwanenburg A, Pilz K et al (2017) A comparative study of machine learning methods for time-to-event survival data for radiomics risk modelling. Sci Rep 7:13206. https://doi.org/10.1038/s41598-017-13448-3
Vallières M, Kay-Rivest E, Perrin LJ et al (2017) Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci Rep 7:10117. https://doi.org/10.1038/s41598-017-10371-5
Sun R, Limkin EJ, Vakalopoulou M et al (2018) A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol 19:1180–1191. https://doi.org/10.1016/S1470-2045(18)30413-3
Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJWL (2015) Machine learning methods for quantitative radiomic biomarkers. Sci Rep 5:13087. https://doi.org/10.1038/srep13087
Guyon I, Elisseeff A (2003) An introduction to variable and feature selection. J Mach Learn Res 3:1157–1182
Brown G, Pocock A, Zhao M-J, Luján M (2012) Conditional likelihood maximisation: a unifying framework for information theoretic feature selection. J Mach Learn Res 13:27–66
Kalousis A, Prados J, Hilario M (2006) Stability of feature selection algorithms: a study on high-dimensional spaces. Knowl Inf Syst 12:95–116. https://doi.org/10.1007/s10115-006-0040-8
Haury A-C, Gestraud P, Vert J-P (2011) The influence of feature selection methods on accuracy, stability and interpretability of molecular signatures. PLoS One 6:e28210. https://doi.org/10.1371/journal.pone.0028210
Mazurowski MA, Habas PA, Zurada JM, Lo JY, Baker JA, Tourassi GD (2008) Training neural network classifiers for medical decision making: the effects of imbalanced datasets on classification performance. Neural Netw Off J Int Neural Netw Soc 21:427–436. https://doi.org/10.1016/j.neunet.2007.12.031
van Smeden M, Moons KG, de Groot JA et al (2019) Sample size for binary logistic prediction models: beyond events per variable criteria. Stat Methods Med Res 28:2455–2474. https://doi.org/10.1177/0962280218784726
Olson RS, La Cava W, Mustahsan Z, Varik A, Moore JH (2018) Data-driven advice for applying machine learning to bioinformatics problems. Pac Symp Biocomput 23:192–203
Dankers FJWM, Traverso A, Wee L, van Kuijk SMJ (2019) Prediction modeling methodology. In: Kubben P, Dumontier M, Dekker A (eds) Fundamentals of clinical data science. Springer, Cham
Vickers AJ, Elkin EB (2006) Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making 26:565–574. https://doi.org/10.1177/0272989X06295361
Vickers AJ, van Calster B, Steyerberg EW (2019) A simple, step-by-step guide to interpreting decision curve analysis. Diagn Progn Res 3:18. https://doi.org/10.1186/s41512-019-0064-7
de Sitter A, Visser M, Brouwer I et al (2020) Facing privacy in neuroimaging: removing facial features degrades performance of image analysis methods. Eur Radiol 30:1062–1074. https://doi.org/10.1007/s00330-019-06459-3
Mongan J, Moy L, Kahn CE (2020) Checklist for Artificial Intelligence in Medical Imaging (CLAIM): a guide for authors and reviewers. Radiology Artificial Intelligence 2:e200029. https://doi.org/10.1148/ryai.2020200029
Luo W, Phung D, Tran T et al (2016) Guidelines for developing and reporting machine learning predictive models in biomedical research: a multidisciplinary view. J Med Internet Res 18:e323. https://doi.org/10.2196/jmir.5870
Collins GS, Reitsma JB, Altman DG, Moons KGM (2015) Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. Ann Intern Med 162:55–63. https://doi.org/10.7326/M14-0697
Collins GS, Moons KGM (2019) Reporting of artificial intelligence prediction models. Lancet 393:1577–1579. https://doi.org/10.1016/S0140-6736(19)30037-6
Funding
The authors state that this work has not received any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Burak Kocak, MD.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No statistical methods were necessary for this paper.
Informed consent
Not required.
Ethical approval
Not required.
Methodology
• Review Article
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kocak, B., Kus, E.A. & Kilickesmez, O. How to read and review papers on machine learning and artificial intelligence in radiology: a survival guide to key methodological concepts. Eur Radiol 31, 1819–1830 (2021). https://doi.org/10.1007/s00330-020-07324-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07324-4