Abstract
Objectives
To apply deep learning algorithms using a conventional convolutional neural network (CNN) and a recurrent CNN to differentiate three breast cancer molecular subtypes on MRI.
Methods
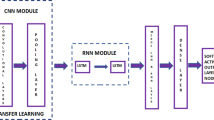
A total of 244 patients were analyzed, 99 in training dataset scanned at 1.5 T and 83 in testing-1 and 62 in testing-2 scanned at 3 T. Patients were classified into 3 subtypes based on hormonal receptor (HR) and HER2 receptor: (HR+/HER2−), HER2+, and triple negative (TN). Only images acquired in the DCE sequence were used in the analysis. The smallest bounding box covering tumor ROI was used as the input for deep learning to develop the model in the training dataset, by using a conventional CNN and the convolutional long short-term memory (CLSTM). Then, transfer learning was applied to re-tune the model using testing-1(2) and evaluated in testing-2(1).
Results
In the training dataset, the mean accuracy evaluated using tenfold cross-validation was higher by using CLSTM (0.91) than by using CNN (0.79). When the developed model was applied to the independent testing datasets, the accuracy was 0.4–0.5. With transfer learning by re-tuning parameters in testing-1, the mean accuracy reached 0.91 by CNN and 0.83 by CLSTM, and improved accuracy in testing-2 from 0.47 to 0.78 by CNN and from 0.39 to 0.74 by CLSTM. Overall, transfer learning could improve the classification accuracy by greater than 30%.
Conclusions
The recurrent network using CLSTM could track changes in signal intensity during DCE acquisition, and achieved a higher accuracy compared with conventional CNN during training. For datasets acquired using different settings, transfer learning can be applied to re-tune the model and improve accuracy.
Key Points
• Deep learning can be applied to differentiate breast cancer molecular subtypes.
• The recurrent neural network using CLSTM could track the change of signal intensity in DCE images, and achieved a higher accuracy compared with conventional CNN during training.
• For datasets acquired using different scanners with different imaging protocols, transfer learning provided an efficient method to re-tune the classification model and improve accuracy.





Similar content being viewed by others
Abbreviations
- ADC:
-
Apparent diffusion coefficient
- AUC:
-
Area under the ROC curve
- CAD:
-
Computer-aided diagnosis
- CLSTM:
-
Convolutional long short-term memory
- CNN:
-
Convolutional neural network
- DCE-MRI:
-
Dynamic contrast-enhanced magnetic resonance imaging
- FCM:
-
Fuzzy-C-means
- GLCM:
-
Gray level co-occurrence matrix
- HR:
-
Hormonal receptor
- LSTM:
-
Long short-term memory
- RNN:
-
Recurrent neural network
- ROC:
-
Receiver operating characteristic
- ROI:
-
Region of interest
- SE:
-
Signal enhancement
- SVM:
-
Support vector machine
- TN:
-
Triple negative
References
Sandhu R, Parker JS, Jones WD, Livasy CA, Coleman WB (2010) Microarray-based gene expression profiling for molecular classification of breast cancer and identification of new targets for therapy. Lab Med 41:364–372
Houssami N, Turner RM, Morrow M (2017) Meta-analysis of pre-operative magnetic resonance imaging (MRI) and surgical treatment for breast cancer. Breast Cancer Res Treat 165(2):273–283
Agner SC, Rosen MA, Englander S et al (2014) Computerized image analysis for identifying triple-negative breast cancers and differentiating them from other molecular subtypes of breast cancer on dynamic contrast-enhanced MR images: a feasibility study. Radiology. 272:91–99
Chang R-F, Chen H-H, Chang Y-C, Huang C-S, Chen J-H, Lo C-M (2016) Quantification of breast tumor heterogeneity for ER status, HER2 status, and TN molecular subtype evaluation on DCE-MRI. Magn Reson Imaging 34:809–819
Sutton EJ, Dashevsky BZ, Oh JH et al (2016) Breast cancer molecular subtype classifier that incorporates MRI features. J Magn Reson Imaging 44:122–129
Li H, Zhu Y, Burnside ES et al (2016) Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer 2:16012
Fan M, Li H, Wang S, Zheng B, Zhang J, Li L (2017) Radiomic analysis reveals DCE-MRI features for prediction of molecular subtypes of breast cancer. PLoS One 12:e0171683
Braman NM, Etesami M, Prasanna P et al (2017) Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res 19:57
Ma W, Zhao Y, Ji Y et al (2019) Breast cancer molecular subtype prediction by mammographic radiomic features. Acad Radiol 26:196–201
Shi L, Zhang Y, Nie K et al (2019) Machine learning for prediction of chemoradiation therapy response in rectal cancer using pre-treatment and mid-radiation multi-parametric MRI. Magn Reson Imaging 61:33–40
Truhn D, Schrading S, Haarburger C, Schneider H, Merhof D, Kuhl C (2019) Radiomic versus convolutional neural networks analysis for classification of contrast-enhancing lesions at multiparametric breast MRI. Radiology. 290:290–297
Antropova N, Huynh B, Li H, Giger ML (2019) Breast lesion classification based on dynamic contrast-enhanced magnetic resonance images sequences with long short-term memory networks. J Med Imaging (Bellingham) 6(1):011002
Zhou J, Zhang Y, Chang KT et al (2020) Diagnosis of benign and malignant breast lesions on DCE-MRI by using radiomics and deep learning with consideration of peritumor tissue. J Magn Reson Imaging 51(3):798–809
Zhu Z, Albadawy E, Saha A, Zhang J, Harowicz MR, Mazurowski MA (2019) Deep learning for identifying radiogenomic associations in breast cancer. Comput Biol Med 109:85–90
Xie T, Wang Z, Zhao Q et al (2019) Machine learning-based analysis of MR multiparametric radiomics for the subtype classification of breast cancer. Front Oncol 9:505
Ha R, Mutasa S, Karcich J et al (2019) Predicting breast cancer molecular subtype with MRI dataset utilizing convolutional neural network algorithm. J Digit Imaging 32:276–282
Chang P, Grinband J, Weinberg B et al (2018) Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. AJNR Am J Neuroradiol 39(7):1201–1207
He K, Zhang X, Ren S, Sun J (2015) Delving deep into rectifiers: Surpassing human-level performance on ImageNet classification. Proceedings of the IEEE international conference on computer vision, pp 1026–1034
Kingma D, Ba J (2014) Adam: a method for stochastic optimization. arXiv preprint arXiv:14126980
Shi X, Chen Z, Wang H, Yeung D-Y, Wong W-K, Woo W-C (2015) Convolutional LSTM network: a machine learning approach for precipitation nowcasting. NIPS’15: Proceedings of the 28th International Conference on Neural Information Processing Systems, pp 802–810
Michael KY, Ma J, Fisher J, Kreisberg JF, Raphael BJ, Ideker T (2018) Visible machine learning for biomedicine. Cell. 173:1562–1565
Antropova N, Abe H, Giger ML (2018) Use of clinical MRI maximum intensity projections for improved breast lesion classification with deep convolutional neural networks. J Med Imaging 5:014503
Haarburger C, Langenberg P, Truhn D et al (2018) Transfer learning for breast cancer malignancy classification based on dynamic contrast- enhanced MR images. In: Maier A, Deserno T, Handels H, Maier-Hein K, Palm C, Tolxdorff T (eds) Bild-verarbeitung für die Medizin. Springer, Berlin, pp 216–221
Lang N, Zhang Y, Zhang E et al (2019) Differentiation of spinal metastases originated from lung and other cancers using radiomics and deep learning based on DCE-MRI. Magn Reson Imaging 64:4–12
Hochreiter S, Schmidhuber J (1997) Long short-term memory. Neural Comput 9:1735–1780
Couture HD, Williams LA, Geradts J et al (2018) Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype. NPJ Breast Cancer 4:30
Jaber MI, Song B, Taylor C et al (2020) A deep learning image-based intrinsic molecular subtype classifier of breast tumors reveals tumor heterogeneity that may affect survival. Breast Cancer Res 22:12
Nishio M, Sugiyama O, Yakami M et al (2018) Computer-aided diagnosis of lung nodule classification between benign nodule, primary lung cancer, and metastatic lung cancer at different image size using deep convolutional neural network with transfer learning. PLoS One 13:e0200721
Yuan Y, Qin W, Buyyounouski M et al (2019) Prostate cancer classification with multiparametric MRI transfer learning model. Med Phys 46:756–765
Byra M, Galperin M, Ojeda-Fournier H et al (2019) Breast mass classification in sonography with transfer learning using a deep convolutional neural network and color conversion. Med Phys 46:746–755
Samala RK, Chan H-P, Hadjiiski L, Helvie MA, Richter CD, Cha KH (2019) Breast cancer diagnosis in digital breast tomosynthesis: effects of training sample size on multi-stage transfer learning using deep neural nets. IEEE Trans Med Imaging 38:686–696
Ortiz-Ramón R, Larroza A, Ruiz-España S, Arana E, Moratal D (2018) Classifying brain metastases by their primary site of origin using a radiomics approach based on texture analysis: a feasibility study. Eur Radiol 28:4514–4523
Funding
This study has received funding by NIH/NCI R01 CA127927, R21 CA208938.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Min-Ying Su, PhD, Professor of Radiological Sciences at University of California, Irvine.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic study
• performed at two institutions
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Drs. Jeon-Hor Chen, Meihao Wang and Min-Ying Su contributed equally to this paper.
Rights and permissions
About this article
Cite this article
Zhang, Y., Chen, JH., Lin, Y. et al. Prediction of breast cancer molecular subtypes on DCE-MRI using convolutional neural network with transfer learning between two centers. Eur Radiol 31, 2559–2567 (2021). https://doi.org/10.1007/s00330-020-07274-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07274-x