Abstract
Objective
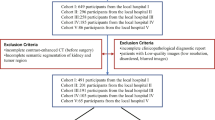
To determine the possible influence of segmentation margin on each step (feature reproducibility, selection, and classification) of the machine learning (ML)-based high-dimensional quantitative computed tomography (CT) texture analysis (qCT-TA) of renal clear cell carcinomas (RcCCs).
Materials and methods
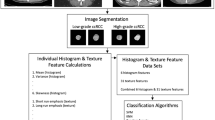
For this retrospective study, 47 patients with RcCC were included from a public database. Two segmentations were obtained by two radiologists for each tumour: (i) contour-focused and (ii) margin shrinkage of 2 mm. Texture features were extracted from original, filtered, and transformed CT images. Feature selection was done using a correlation-based algorithm. The ML classifier was k-nearest neighbours. Classifications were performed with and without using synthetic minority over-sampling technique. Reference standard was nuclear grade (low versus high). Intraclass correlation coefficient (ICC), Pearson’s correlation coefficient, Wilcoxon signed-ranks test, and McNemar’s test were used in the analysis.
Results
The segmentation with margin shrinkage of 2 mm (772 of 828; 93.2%) yielded more texture features with excellent reproducibility (ICC ≥ 0.9) than contour-focused segmentation (714 of 828; 86.2%), p < 0.0001. The feature selection algorithms resulted in different feature subsets for two segmentation datasets with only one common feature. All ML-based models based on contour-focused segmentation (area under the curve [AUC] range, 0.865–0.984) performed better than those with margin shrinkage of 2 mm (AUC range, 0.745–0.887), p < 0.05.
Conclusions
Each step of the ML-based high-dimensional qCT-TA was susceptible to a slight change of 2 mm in segmentation margin. Despite yielding fewer features with excellent reproducibility, use of the contour-focused segmentation provided better classification performance for distinguishing nuclear grade.
Key Points
• Each step of a machine learning (ML)-based high-dimensional quantitative computed tomography texture analysis (qCT-TA) is sensitive to even a slight change of 2 mm in segmentation margin.
• Despite yielding fewer texture features with excellent reproducibility, performing the segmentation focusing on the outermost boundary of the tumours provides better classification performance in ML-based qCT-TA of renal clear cell carcinomas for distinguishing nuclear grade.
• Findings of an ML-based high-dimensional qCT-TA may not be reproducible in clinical practice even using the same feature selection algorithm and ML classifier unless the possible influence of the segmentation margin is considered.






Similar content being viewed by others
Abbreviations
- AUC:
-
Area under the curve
- CE-CT:
-
Contrast-enhanced computed tomography
- CT:
-
Computed tomography
- ICC:
-
Intraclass correlation coefficient
- k-NN:
-
k-nearest neighbours
- LoG:
-
Laplacian of Gaussian
- NN:
-
Nearest neighbours
- qCT-TA:
-
Quantitative computed tomography texture analysis
- qTA:
-
Quantitative texture analysis
- RCC:
-
Renal cell carcinoma
- RcCC:
-
Renal clear cell carcinoma
- TCGA-KIRC:
-
The Cancer Genome Atlas-Kidney Renal Clear Cell Carcinoma
- WEKA:
-
Waikato environment for knowledge analysis
References
Lubner MG, Smith AD, Sandrasegaran K, Sahani DV, Pickhardt PJ (2017) CT texture analysis: definitions, applications, biologic correlates, and challenges. RadioGraphics 37:1483–1503. https://doi.org/10.1148/rg.2017170056
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577. https://doi.org/10.1148/radiol.2015151169
Kocak B, Yardimci AH, Bektas CT et al (2018) Textural differences between renal cell carcinoma subtypes: machine learning-based quantitative computed tomography texture analysis with independent external validation. Eur J Radiol 107:149–157. https://doi.org/10.1016/j.ejrad.2018.08.014
Bektas CT, Kocak B, Yardimci AH et al (2018) Clear cell renal cell carcinoma: machine learning-based quantitative computed tomography texture analysis for prediction of Fuhrman nuclear grade. Eur Radiol. https://doi.org/10.1007/s00330-018-5698-2
Berenguer R, Pastor-Juan MDR, Canales-Vázquez J et al (2018) Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology 288:407–415. https://doi.org/10.1148/radiol.2018172361
Shafiq-ul-Hassan M, Zhang GG, Latifi K et al (2017) Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys 44:1050–1062. https://doi.org/10.1002/mp.12123
Mackin D, Fave X, Zhang L et al (2015) Measuring computed tomography scanner variability of radiomics features. Invest Radiol 50:757–765. https://doi.org/10.1097/RLI.0000000000000180
Balagurunathan Y, Gu Y, Wang H et al (2014) Reproducibility and prognosis of quantitative features extracted from CT images. Transl Oncol 7:72–87
Balagurunathan Y, Kumar V, Gu Y et al (2014) Test-retest reproducibility analysis of lung CT image features. J Digit Imaging 27:805–823. https://doi.org/10.1007/s10278-014-9716-x
Hunter LA, Krafft S, Stingo F et al (2013) High quality machine-robust image features: identification in nonsmall cell lung cancer computed tomography images. Med Phys 40:121916. https://doi.org/10.1118/1.4829514
Leijenaar RT, Carvalho S, Velazquez ER et al (2013) Stability of FDG-PET radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol 52:1391–1397. https://doi.org/10.3109/0284186X.2013.812798
Echegaray S, Gevaert O, Shah R et al (2015) Core samples for radiomics features that are insensitive to tumor segmentation: method and pilot study using CT images of hepatocellular carcinoma. J Med Imaging (Bellingham) 2:041011. https://doi.org/10.1117/1.JMI.2.4.041011
Parmar C, Rios Velazquez E, Leijenaar R et al (2014) Robust radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One 9:e102107. https://doi.org/10.1371/journal.pone.0102107
Fave X, Zhang L, Yang J et al (2016) Impact of image preprocessing on the volume dependence and prognostic potential of radiomics features in non-small cell lung cancer. Transl Cancer Res 5:349–363. https://doi.org/10.21037/8709
Hu P, Wang J, Zhong H et al (2016) Reproducibility with repeat CT in radiomics study for rectal cancer. Oncotarget 7:71440–71446. https://doi.org/10.18632/oncotarget.12199
Kim H, Park CM, Lee M et al (2016) Impact of reconstruction algorithms on CT radiomic features of pulmonary tumors: analysis of intra- and inter-reader variability and inter-reconstruction algorithm variability. PLoS One 11:e0164924. https://doi.org/10.1371/journal.pone.0164924
Akin O, Elnajjar P, Heller M, et al (2016) Radiology Data from The Cancer Genome Atlas Kidney Renal Clear Cell Carcinoma [TCGA-KIRC] collection. The Cancer Imaging Archive. https://wiki.cancerimagingarchive.net/display/Public/TCGA-KIRC#329b9f4f31cf4586831934c19c4f10f4. Accessed 4 May 2018
Clark K, Vendt B, Smith K et al (2013) The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 26:1045–1057. https://doi.org/10.1007/s10278-013-9622-7
Kocak B, Durmaz ES, Ates E, Ulusan MB (2019) Radiogenomics in clear cell renal cell carcinoma: machine learning–based high-dimensional quantitative CT texture analysis in predicting PBRM1 mutation status. AJR Am J Roentgenol 1–9. https://doi.org/10.2214/AJR.18.20443
Cohan RH, Sherman LS, Korobkin M, Bass JC, Francis IR (1995) Renal masses: assessment of corticomedullary-phase and nephrographic-phase CT scans. Radiology 196:445–451. https://doi.org/10.1148/radiology.196.2.7617859
van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107. https://doi.org/10.1158/0008-5472.CAN-17-0339
Koo TK, Li MY (2016) A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med 15:155–163. https://doi.org/10.1016/j.jcm.2016.02.012
Kohavi R, John GH (1997) Wrappers for feature subset selection. Artif Intell 97:273–324. https://doi.org/10.1016/S0004-3702(97)00043-X
Bermejo P, Gamez JA, Puerta JM (2011) Improving incremental wrapper-based subset selection via replacement and early stopping. Intern J Pattern Recognit Artif Intell 25:605–625. https://doi.org/10.1142/S0218001411008804
Dormann CF, Elith J, Bacher S et al (2013) Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography (Cop) 36:27–46. https://doi.org/10.1111/j.1600-0587.2012.07348.x
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP (2002) SMOTE: synthetic minority over-sampling technique. J Artif Intell Res 16:321–357. https://doi.org/10.1613/JAIR.953
Demšar J (2006) Statistical comparisons of classifiers over multiple data sets. J Mach Learn Res 7:1–30
Yan L, Liu Z, Wang G et al (2015) Angiomyolipoma with minimal fat. Acad Radiol 22:1115–1121. https://doi.org/10.1016/j.acra.2015.04.004
Hodgdon T, McInnes MD, Schieda N, Flood TA, Lamb L, Thornhill RE (2015) Can quantitative CT texture analysis be used to differentiate fat-poor renal angiomyolipoma from renal cell carcinoma on unenhanced CT images? Radiology 276:787–796. https://doi.org/10.1148/radiol.2015142215
Klatte T, Patard JJ, de Martino M et al (2008) Tumor size does not predict risk of metastatic disease or prognosis of small renal cell carcinomas. J Urol 179:1719–1726. https://doi.org/10.1016/j.juro.2008.01.018
Frank I, Blute ML, Cheville JC, Lohse CM, Weaver AL, Zincke H (2002) An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade and necrosis: the SSIGN score. J Urol 168:2395–2400. https://doi.org/10.1097/01.ju.0000035885.91935.d5
Zisman A, Pantuck AJ, Dorey F et al (2002) Mathematical model to predict individual survival for patients with renal cell carcinoma. J Clin Oncol 20:1368–1374. https://doi.org/10.1200/JCO.2002.20.5.1368
Marconi L, Dabestani S, Lam TB et al (2016) Systematic review and meta-analysis of diagnostic accuracy of percutaneous renal tumour biopsy. Eur Urol 69:660–673. https://doi.org/10.1016/j.eururo.2015.07.072
Jewett MAS, Mattar K, Basiuk J et al (2011) Active surveillance of small renal masses: progression patterns of early stage kidney cancer. Eur Urol 60:39–44. https://doi.org/10.1016/j.eururo.2011.03.030
Abou Youssif T, Tanguay S (2009) Natural history and management of small renal masses. Curr Oncol 16(Suppl 1):S2–S7
Ng F, Kozarski R, Ganeshan B, Goh V (2013) Assessment of tumor heterogeneity by CT texture analysis: can the largest cross-sectional area be used as an alternative to whole tumor analysis? Eur J Radiol 82:342–348. https://doi.org/10.1016/j.ejrad.2012.10.023
Schieda N, Lim RS, Krishna S, McInnes MDF, Flood TA, Thornhill RE (2018) Diagnostic accuracy of unenhanced CT analysis to differentiate low-grade from high-grade chromophobe renal cell carcinoma. Am J Roentgenol:1–9. https://doi.org/10.2214/AJR.17.18874
Collewet G, Strzelecki M, Mariette F (2004) Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn Reson Imaging 22:81–91. https://doi.org/10.1016/j.mri.2003.09.001
Acknowledgements
The abstract of this study has also been submitted to the European Congress of Radiology (ECR) 2019 (Control Number 19-P-339-ECR). The authors acknowledge that they have previously used this public database (The Cancer Genome Atlas-Kidney Renal Clear Cell Carcinoma [TCGA-KIRC]) in different context.
Funding
The authors state that this work has not received any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Burak Kocak, MD.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors (Burak Kocak, MD) has significant statistical expertise.
Informed consent
Written informed consent was not required for this study because all patients’ data included in this study are publicly and freely available for scientific purposes (The Cancer Genome Atlas-Kidney Renal Clear Cell Carcinoma [TCGA-KIRC]).
Ethical approval
Institutional Review Board approval was not required because all patients’ data included in this study are publicly and freely available for scientific purposes (The Cancer Genome Atlas-Kidney Renal Clear Cell Carcinoma [TCGA-KIRC]).
Study subjects or cohorts overlap
The authors acknowledge that they have previously used this public database (The Cancer Genome Atlas-Kidney Renal Clear Cell Carcinoma [TCGA-KIRC]) in different context.
Methodology
• retrospective
• experimental
• performed using a publicly and freely available database
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 48 kb)
Rights and permissions
About this article
Cite this article
Kocak, B., Ates, E., Durmaz, E.S. et al. Influence of segmentation margin on machine learning–based high-dimensional quantitative CT texture analysis: a reproducibility study on renal clear cell carcinomas. Eur Radiol 29, 4765–4775 (2019). https://doi.org/10.1007/s00330-019-6003-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-019-6003-8