Abstract
Objectives
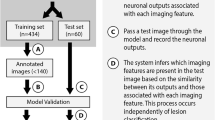
To develop and validate a proof-of-concept convolutional neural network (CNN)–based deep learning system (DLS) that classifies common hepatic lesions on multi-phasic MRI.
Methods
A custom CNN was engineered by iteratively optimizing the network architecture and training cases, finally consisting of three convolutional layers with associated rectified linear units, two maximum pooling layers, and two fully connected layers. Four hundred ninety-four hepatic lesions with typical imaging features from six categories were utilized, divided into training (n = 434) and test (n = 60) sets. Established augmentation techniques were used to generate 43,400 training samples. An Adam optimizer was used for training. Monte Carlo cross-validation was performed. After model engineering was finalized, classification accuracy for the final CNN was compared with two board-certified radiologists on an identical unseen test set.
Results
The DLS demonstrated a 92% accuracy, a 92% sensitivity (Sn), and a 98% specificity (Sp). Test set performance in a single run of random unseen cases showed an average 90% Sn and 98% Sp. The average Sn/Sp on these same cases for radiologists was 82.5%/96.5%. Results showed a 90% Sn for classifying hepatocellular carcinoma (HCC) compared to 60%/70% for radiologists. For HCC classification, the true positive and false positive rates were 93.5% and 1.6%, respectively, with a receiver operating characteristic area under the curve of 0.992. Computation time per lesion was 5.6 ms.
Conclusion
This preliminary deep learning study demonstrated feasibility for classifying lesions with typical imaging features from six common hepatic lesion types, motivating future studies with larger multi-institutional datasets and more complex imaging appearances.
Key Points
• Deep learning demonstrates high performance in the classification of liver lesions on volumetric multi-phasic MRI, showing potential as an eventual decision-support tool for radiologists.
• Demonstrating a classification runtime of a few milliseconds per lesion, a deep learning system could be incorporated into the clinical workflow in a time-efficient manner.




Similar content being viewed by others
Abbreviations
- CNN:
-
Convolutional neural network
- CRC:
-
Colorectal carcinoma
- DL:
-
Deep learning
- DLS:
-
Deep learning system
- FNH:
-
Focal nodular hyperplasia
- HCC:
-
Hepatocellular carcinoma
- ICC:
-
Intrahepatic cholangiocarcinoma
- LI-RADS:
-
Liver Imaging Reporting and Data System
- PACS:
-
Picture archiving and communication system
- Sn:
-
Sensitivity
- Sp:
-
Specificity
References
El–Serag HB, Rudolph KL (2007) Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 132:2557–2576
Wang H, Naghavi M, Allen C et al (2016) Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388:1459–1544
Siegel RL, Miller KD, Jemal A (2016) Cancer statistics, 2016. CA Cancer J Clin 66:7–30
Mitchell DG, Bruix J, Sherman M, Sirlin CB (2015) LI-RADS (Liver Imaging Reporting and Data System): summary, discussion, and consensus of the LI-RADS Management Working Group and future directions. Hepatology 61:1056–1065
Yasaka K, Akai H, Abe O, Kiryu S (2018) Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: a preliminary study. Radiology 286:887–896. https://doi.org/10.1148/radiol.2017170706
Grewal M, Srivastava MM, Kumar P, Varadarajan S (2018) RADnet: radiologist level accuracy using deep learning for hemorrhage detection in CT scans2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pp 281–284
Klöppel S, Stonnington CM, Barnes J et al (2008) Accuracy of dementia diagnosis—a direct comparison between radiologists and a computerized method. Brain 131:2969–2974
Greenspan H, Van Ginneken B, Summers RM (2016) Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans Med Imaging 35:1153–1159
Shiraishi J, Sugimoto K, Moriyasu F, Kamiyama N (2008) Computer-aided diagnosis for the classification of focal liver lesions by use of contrast-enhanced ultrasonography. Med Phys 35:1734–1746
Sugimoto K, Shiraishi J, Moriyasu F, Doi K (2010) Computer-aided diagnosis for contrast-enhanced ultrasound in the liver. World J Radiol 2:215
Hwang YN, Lee JH, Kim GY, Jiang YY, Kim SM (2015) Classification of focal liver lesions on ultrasound images by extracting hybrid textural features and using an artificial neural network. Biomed Mater Eng 26:S1599–S1611
Virmani J, Kumar V, Kalra N, Khandelwa N (2013) PCA-SVM based CAD system for focal liver lesions using B-mode ultrasound images. Def Sci J 63:478
Acharya UR, Koh JEW, Hagiwara Y et al (2018) Automated diagnosis of focal liver lesions using bidirectional empirical mode decomposition features. Comput Biol Med 94:11–18
Rajpurkar P, Irvin J, Ball RL et al (2018) Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med 15:e1002686. https://doi.org/10.1371/journal.pmed.1002686
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. pp 1097–1105
Chollet F (2015) Keras. https://keras.io/. Accessed 15 Oct 2018
Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:150203167
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. arXiv preprint arXiv:14126980
Chapiro J, Lin M, Duran R, Schernthaner RE, Geschwind J-F (2015) Assessing tumor response after loco-regional liver cancer therapies: the role of 3D MRI. Expert Rev Anticancer Ther 15:199
Chapiro J, Wood LD, Lin M et al (2014) Radiologic-pathologic analysis of contrast-enhanced and diffusion-weighted MR imaging in patients with HCC after TACE: diagnostic accuracy of 3D quantitative image analysis. Radiology 273:746–758
Barth B, Donati O, Fischer M et al (2016) Reliability, validity, and reader acceptance of LI-RADS-an in-depth analysis. Acad Radiol 23:1145
Bashir M, Huang R, Mayes N et al (2015) Concordance of hypervascular liver nodule characterization between the organ procurement and transplant network and liver imaging reporting and data system classifications. J Magn Reson Imaging 42:305
Davenport MS, Khalatbari S, Liu PS et al (2014) Repeatability of diagnostic features and scoring systems for hepatocellular carcinoma by using MR imaging. Radiology 272:132
Ehman EC, Behr SC, Umetsu SE et al (2016) Rate of observation and inter-observer agreement for LI-RADS major features at CT and MRI in 184 pathology proven hepatocellular carcinomas. Abdom Radiol (NY) 41:963–969
Fowler KJ, Tang A, Santillan C et al (2018) Interreader reliability of LI-RADS version 2014 algorithm and imaging features for diagnosis of hepatocellular carcinoma: a large international multireader study. Radiology 286:173–185
Liu W, Qin J, Guo R et al (2017) Accuracy of the diagnostic evaluation of hepatocellular carcinoma with LI-RADS. Acta Radiol. https://doi.org/10.1177/0284185117716700:284185117716700
Sirlin CB, Kielar AZ, Tang A, Bashir MR (2018) LI-RADS: a glimpse into the future. Abdom Radiol (NY) 43:231–236
Funding
BL and CW received funding from the Radiological Society of North America (RSNA Research Resident Grant no. RR1731). JD, JC, ML, and CW received funding from the National Institutes of Health (NIH/NCI R01 CA206180).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Brian Letzen.
Conflict of interest
The authors of this manuscript declare relationships with the following companies: JW: Bracco Diagnostics, Siemens AG; ML: Pro Medicus Limited; JC Koninklijke Philips, Guerbet SA, Eisai Co.
Statistics and biometry
One of the authors has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• experimental
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 21.4 kb)
Rights and permissions
About this article
Cite this article
Hamm, C.A., Wang, C.J., Savic, L.J. et al. Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol 29, 3338–3347 (2019). https://doi.org/10.1007/s00330-019-06205-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-019-06205-9