Abstract
Key message
OsVP1 and Sdr4 play an important role in regulating seed dormancy that involved in multiple metabolism and regulatory pathways.
Abstract
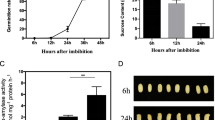
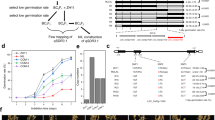
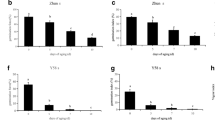
Seed dormancy and germination are critical agricultural traits influencing rice grain yield. Although there are some genes have identified previously, the comprehensive understanding based on transcriptome is still deficient. In this study, we generated mutants of two representative regulators of seed germination, Oryza sativa Viviparous1 (OsVP1) and Seed dormancy 4 (Sdr4), by CRISPR/Cas9 approach and named them cr-osvp1 and cr-sdr4. The weakened dormancy of mutants indicated that the functions of OsVP1 and Sdr4 are required for normal early seed dormancy. There were 4157 and 8285 differentially expressed genes (DEGs) were identified in cr-osvp1 vs. NIP and cr-sdr4 vs. NIP groups, respectively, with a large number of overlapped DEGs between two groups. The gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of common DEGs in two groups showed that genes related to carbohydrate metabolic, nucleoside metabolic, amylase activity and plant hormone signal transduction were involved in the dormancy regulation. These results suggest that OsVP1 and Sdr4 play an important role in regulating seed dormancy by multiple metabolism and regulatory pathways. The systematic analysis of the transcriptional level changes provides theoretical basis for the research of seed dormancy and germination in rice.







Similar content being viewed by others
Data availability
The datasets generated and/or analyzed in the course of this study are available from the corresponding authors on reasonable request.
References
Chen W, Wang W, Lyu Y, Wu Y, Huang P, Hu S, Wei X, Jiao G, Sheng Z, Tang S, Shao G, Luo J (2021) OsVP1 activates Sdr4 expression to control rice seed dormancy via the ABA signaling pathway. Crop J 9(1):68–78. https://doi.org/10.1016/j.cj.2020.06.005
Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, Koshiba T, Sheen J (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14(11):2723–2743. https://doi.org/10.1105/tpc.006494
Corbineau F, Xia Q, Bailly C, El-Maarouf-Bouteau H (2014) Ethylene, a key factor in the regulation of seed dormancy. Front Plant Sci 5:539. https://doi.org/10.3389/fpls.2014.00539
Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61:651–679. https://doi.org/10.1146/annurev-arplant-042809-112122
Damaris RN, Lin Z, Yang P, He D (2019) The rice alpha-amylase, conserved regulator of seed Maturation and germination. Int J Mol Sci 20(2):450. https://doi.org/10.3390/ijms20020450
Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell 127(7):1309–1321. https://doi.org/10.1016/j.cell.2006.12.006
Du L, Xu F, Fang J, Gao S, Tang J, Fang S, Wang H, Tong H, Zhang F, Chu J, Wang G, Chu C (2018) Endosperm sugar accumulation caused by mutation of PHS8/ISA1 leads to pre-harvest sprouting in rice. Plant J 95(3):545–556. https://doi.org/10.1111/tpj.13970
Eastwood D, Tavener RJ, Laidman DL (1969) Sequential action of cytokinin and gibberellic acid in wheat aleurone tissue. Nature 221(5187):1267. https://doi.org/10.1038/2211267a0
El-Maarouf-Bouteau H, Sajjad Y, Bazin J, Langlade N, Cristescu SM, Balzergue S, Baudouin E, Bailly C (2015) Reactive oxygen species, abscisic acid and ethylene interact to regulate sunflower seed germination. Plant Cell Environ 38(2):364–374. https://doi.org/10.1111/pce.12371
Fang J, Chu C (2008) Abscisic acid and the pre-harvest sprouting in cereals. Plant Signal Behav 3(12):1046–1048. https://doi.org/10.4161/psb.3.12.6606
Finkelstein RR, Lynch TJ (2000) Abscisic acid inhibition of radicle emergence but not seedling growth is suppressed by sugars. Plant Physiol 122(4):1179–1186. https://doi.org/10.1104/pp.122.4.1179
Fujii H, Zhu JK (2009) Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA 106(20):8380–8385. https://doi.org/10.1073/pnas.0903144106
Fujii H, Verslues PE, Zhu JK (2007) Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19(2):485–494. https://doi.org/10.1105/tpc.106.048538
Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2006) Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA 103(6):1988–1993. https://doi.org/10.1073/pnas.0505667103
Garciarrubio A, Covarrubias L (1997) Abscisic acid inhibits germination of mature Arabidopsis seeds by limiting the availability of energy and nutrients. Planta 203(2):182–187. https://doi.org/10.1007/s004250050180
Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4(10):1251–1261. https://doi.org/10.1105/tpc.4.10.1251
Graeber K, Nakabayashi K, Miatton E, Leubner-Metzger G, Soppe WJ (2012) Molecular mechanisms of seed dormancy. Plant Cell Environ 35(10):1769–1786. https://doi.org/10.1111/j.1365-3040.2012.02542.x
Hattori T, Terada T, Hamasuna ST (1994) Sequence and functional analyses of the rice gene homologous to the maize Vp1. Plant Mol Biol 24(5):805–810. https://doi.org/10.1007/BF00029862
He D, Yang P (2013) Proteomics of rice seed germination. Front Plant Sci 4:246. https://doi.org/10.3389/fpls.2013.00246
He D, Han C, Yao J, Shen S, Yang P (2011) Constructing the metabolic and regulatory pathways in germinating rice seeds through proteomic approach. Proteomics 11(13):2693–2713. https://doi.org/10.1002/pmic.201000598
He M, Zhu C, Dong K, Zhang T, Cheng Z, Li J, Yan Y (2015) Comparative proteome analysis of embryo and endosperm reveals central differential expression proteins involved in wheat seed germination. BMC Plant Biol 15:97. https://doi.org/10.1186/s12870-015-0471-z
Hisano H, Hoffie RE, Abe F, Munemori H, Matsuura T, Endo M, Mikami M, Nakamura S, Kumlehn J, Sato K (2022) Regulation of germination by targeted mutagenesis of grain dormancy genes in barley. Plant Biotechnol J 20(1):37–46. https://doi.org/10.1111/pbi.13692
Hobo T, Kowyama Y, Hattori T (1999) A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc Natl Acad Sci USA 96(26):15348–15353. https://doi.org/10.1073/pnas.96.26.15348
Holdsworth MJ, Bentsink L, Soppe WJJ (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179(1):33–54. https://doi.org/10.1111/j.1469-8137.2008.02437.x
Hwang I, Sheen J, Müller B (2012) Cytokinin signaling networks. Annu Rev Plant Biol 63:353–380. https://doi.org/10.1146/annurev-arplant-042811-105503
Itoh H, Shimada A, Ueguchi-Tanaka M, Kamiya N, Hasegawa Y, Ashikari M, Matsuoka M (2005) Overexpression of a GRAS protein lacking the DELLA domain confers altered gibberellin responses in rice. Plant J 44(4):669–679. https://doi.org/10.1111/j.1365-313X.2005.02562.x
Ju L, Jing Y, Shi P, Liu J, Chen J, Yan J, Chu J, Chen KM, Sun J (2019) JAZ proteins modulate seed germination through interaction with ABI5 in bread wheat and Arabidopsis. New Phytol 223(1):246–260. https://doi.org/10.1111/nph.15757
Kieber JJ, Schaller GE (2018) Cytokinin signaling in plant development. Development. https://doi.org/10.1242/dev.149344
Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, Yamamoto A, Hattori T (2005) Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J 44(6):939–949. https://doi.org/10.1111/j.1365-313X.2005.02583.x
Liu X, Zhang H, Zhao Y, Feng Z, Li Q, Yang HQ, Luan S, Li J, He ZH (2013) Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc Natl Acad Sci USA 110(38):15485–15490. https://doi.org/10.1073/pnas.1304651110
Magwa RA, Zhao H, Xing Y (2016) Genome-wide association mapping revealed a diverse genetic basis of seed dormancy across subpopulations in rice (Oryza sativa L.). BMC Genet 17:28. https://doi.org/10.1186/s12863-016-0340-2
Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25(3):295–303. https://doi.org/10.1046/j.1365-313x.2001.00965.x
Murase K, Hirano Y, Sun TP, Hakoshima T, Murase K, Hirano Y, Sun TP, Hakoshima T (2008) Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456(7221):459–463. https://doi.org/10.1038/nature07519
Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, Shinozaki K, Yamaguchi-Shinozaki K (2009) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 50(7):1345–1363. https://doi.org/10.1093/pcp/pcp083
Née G, Kramer K, Nakabayashi K, Yuan B, Xiang Y, Miatton E, Finkemeier I, Soppe WJJ (2017) DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat Commun 8(1):72. https://doi.org/10.1038/s41467-017-00113-6
Pan J, Hu Y, Wang H, Guo Q, Chen Y, Howe GA, Yu D (2020) Molecular mechanism underlying the synergetic effect of jasmonate on abscisic acid signaling during seed germination in arabidopsis. Plant Cell 32(12):3846–3865. https://doi.org/10.1105/tpc.19.00838
Powers SK, Holehouse AS, Korasick DA, Schreiber KH, Clark NM, Jing H, Emenecker R, Han S, Tycksen E, Hwang I, Sozzani R, Jez JM, Pappu RV, Strader LC (2019) Nucleo-cytoplasmic partitioning of ARF proteins controls auxin responses in arabidopsis thaliana. Mol Cell 76(1):177-190.e5. https://doi.org/10.1016/j.molcel.2019.06.044
Rolland F, Baena-Gonzalez E, Sheen J (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57:675–709. https://doi.org/10.1146/annurev.arplant.57.032905.105441
Salehin M, Bagchi R, Estelle M (2015) SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27(1):9–19. https://doi.org/10.1105/tpc.114.133744
Schmittgen TD, Livak KJ, Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3(6):1101–1108. https://doi.org/10.1038/nprot.2008.73
Shu K, Liu XD, Xie Q, He ZH (2016) Two faces of one seed: hormonal regulation of dormancy and germination. Mol Plant 9(1):34–45. https://doi.org/10.1016/j.molp.2015.08.010
Shuai HW, Meng YJ, Luo XF, Chen F, Qi Y, Yang WY, Shu K (2016) The roles of auxin in seed dormancy and germination. Yi Chuan 38(4):314–322
Song S, Wang G, Wu H, Fan X, Liang L, Zhao H, Li S, Hu Y, Liu H, Ayaad M, Xing Y (2020) OsMFT2 is involved in the regulation of ABA signaling-mediated seed germination through interacting with OsbZIP23/66/72 in rice. Plant J 103(2):532–546. https://doi.org/10.1111/tpj.14748
Sugimoto K, Takeuchi Y, Ebana K, Miyao A, Hirochika H, Hara N, Ishiyama K, Kobayashi M, Ban Y, Hattori T, Yano M (2010) Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc Natl Acad Sci USA 107(13):5792–5797. https://doi.org/10.1073/pnas.0911965107
Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448(7154):661–665. https://doi.org/10.1038/nature05960
Varshney V, Majee M (2021) JA Shakes hands with ABA to delay seed germination. Trends Plant Sci 26(8):764–766. https://doi.org/10.1016/j.tplants.2021.05.002
Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Laurière C, Merlot S (2009) Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21(10):3170–3184. https://doi.org/10.1105/tpc.109.069179
Winkler M, Niemeyer M, Hellmuth A, Janitza P, Christ G, Samodelov SL, Wilde V, Majovsky P, Trujillo M, Zurbriggen MD, Hoehenwarter W, Quint M, Calderón Villalobos LIA (2017) Variation in auxin sensing guides AUX/IAA transcriptional repressor ubiquitylation and destruction. Nat Commun 8:15706. https://doi.org/10.1038/ncomms15706
Xiang Y, Nakabayashi K, Ding J, He F, Bentsink L, Soppe WJ (2014) REDUCED DORMANCY5 encodes a protein phosphatase 2C that is required for seed dormancy in Arabidopsis. Plant Cell 26(11):4362–4375. https://doi.org/10.1105/tpc.114.132811
Yang P, Li X, Wang X, Chen H, Chen F, Shen S (2007) Proteomic analysis of rice (Oryza sativa) seeds during germination. Proteomics 7(18):3358–3368. https://doi.org/10.1002/pmic.200700207
Zubo YO, Schaller GE (2020) Role of the cytokinin-activated type-B response regulators in hormone crosstalk. Plants (basel). https://doi.org/10.3390/plants9020166
Funding
This research was financially supported by the National Natural Science Foundation of China (32072528, 31872051), and Shenzhen Technology Research & Development (JCYJ20200109150713553).
Author information
Authors and Affiliations
Contributions
WQC designed and performed most of the experiments; JL provided instructions for the experiments; JLX conceived the project and supervised the experiments; ZXL performed the transcriptome data analysis; WQC analyzed the data and wrote the manuscript; DPL, KC and JLX revised the article.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Kinya Toriyama.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
299_2022_2958_MOESM1_ESM.pdf
Supplementary file1 Table S1. Significant GO terms of DEGs in the biological process category for cr-osvp1 vs. NIP. Table S2. Significant GO terms of DEGs in the cellular component category for cr-osvp1 vs. NIP. Table S3. Significant GO terms of DEGs in molecular function category for cr-osvp1 vs. NIP. Table S4. Significant GO terms of DEGs in the biological process category for cr-sdr4 vs. NIP. Table S5. Significant GO terms of DEGs in the cellular component category for cr-sdr4 vs. NIP. Table S6. Significant GO terms of DEGs in molecular function category for cr-sdr4 vs. NIP. Table S7. Primers of qRT-PCR. Fig. S1 Amino acid sequence alignment. (PDF 317 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, WQ., Liu, DP., Li, ZX. et al. Transcriptome analysis of knockout mutants of rice seed dormancy gene OsVP1 and Sdr4. Plant Cell Rep 42, 309–319 (2023). https://doi.org/10.1007/s00299-022-02958-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-022-02958-8