Abstract
Key message
The DNA fragments transferred among cotton cytoplasmic genomes are highly differentiated. The wild D group cotton species have undergone much greater evolution compared with cultivated AD group.
Abstract
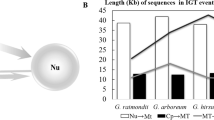
Cotton (Gossypium spp.) is one of the most economically important fiber crops worldwide. Gene transfer, nucleotide evolution, and the codon usage preferences in cytoplasmic genomes are important evolutionary characteristics of high plants. In this study, we analyzed the nucleotide sequence evolution, codon usage, and transfer of cytoplasmic DNA fragments in Gossypium chloroplast (cp) and mitochondrial (mt) genomes, including the A genome group, wild D group, and cultivated AD group of cotton species. Our analyses indicated that the differences in the length of transferred cytoplasmic DNA fragments were not significant in mitochondrial and chloroplast sequences. Analysis of the transfer of tRNAs found that trnQ and nine other tRNA genes were commonly transferred between two different cytoplasmic genomes. The Codon Adaptation Index values showed that Gossypium cp genomes prefer A/T-ending codons. Codon preference selection was higher in the D group than the other two groups. Nucleotide sequence evolution analysis showed that intergenic spacer sequences were more variable than coding regions and nonsynonymous mutations were clearly more common in cp genomes than mt genomes. Evolutionary analysis showed that the substitution rate was much higher in cp genomes than mt genomes. Interestingly, the D group cotton species have undergone much faster evolution compared with cultivated AD groups, possibly due to the selection and domestication of diverse cotton species. Our results demonstrate that gene transfer and differential nucleotide sequence evolution have occurred frequently in cotton cytoplasmic genomes.






Similar content being viewed by others
References
Adams KL, Wendel JF (2004) Exploring the genomic mysteries of polyploidy in cotton. Biol J Linnean Soc 82(4):573–581
Agris PF (2004) Decoding the genome: a modified view. Nucleic Acids Res 32(1):223–238
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Amaral-Zettler LA, Dragone NB, Schell J, Slikas B, Murphy LG, Morrall CE, Zettler ER (2017) Comparative mitochondrial and chloroplast genomics of a genetically distinct form of sargassum contributing to recent "golden tides" in the western Atlantic. Ecol Evol 7(2):516–525
Beasley JO (1940) The origin of American tetraploid Gossypium species. Am Nat 74(752):285–286
Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF (2013) MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol 69(2):313–319
Braukmann T, Kuzmina M, Stefanović S (2013) Plastid genome evolution across the genus Cuscuta (Convolvulaceae): two clades within subgenus grammica exhibit extensive gene loss. J Exp Bot 64(4):977–989
Bright SW, Wood EA, Miflin BJ (1978) The effect of aspartate-derived amino acids (Lysine, Threonine, Methionine) on the growth of excised embryos of wheat and barley. Planta 139(2):113–117
Buschiazzo E, Ritland C, Bohlmann J, Ritland K (2012) Slow but not low: genomic comparisons reveal slower evolutionary rate and higher dN/dS in conifers compared to angiosperms. BMC Evol Biol 12:8
Cai CP, Zhang XY, Niu E, Zhao L, Li N, Wang LM, Ding LY, Guo WZ (2014) GhPSY, a phytoene synthase gene, is related to the red plant phenotype in upland cotton (Gossypium hirsutum L.). Mol Biol Rep 41(8):4941–4952
Cai XY, Liu F, Zhou ZL, Wang XX, Wang CY, Wang YH, Wang KB (2015) Characterization and development of chloroplast microsatellite markers for Gossypium hirsutum, and cross-species amplification in other Gossypium species. Genet Mol Res 14(4):11924–11932
Chen ZW, Grover CE, Li PB, Wang YM, Nie HS, Zhao YP, Wang MY, Liu F, Zhou ZL, Wang XX, Cai XY, Wang KB, Wendel JF, Hua JP (2017a) Molecular evolution of the plastid genome during diversification of the cotton genus. Mol Phylogenet Evol 112:268–276
Chen ZW, Nie HS, Wang YM, Pei HL, Li SS, Zhang LD, Hua JP (2017b) Rapid evolutionary divergence of diploid and allotetraploid Gossypium mitochondrial genomes. BMC Genom 18:876
Collins DW, Jukes TH (1994) Rates of transition and transversion in coding sequences since the human-rodent divergence. Genomics 20(3):386–396
Cronn RC, Small RL, Haselkorn T, Wendel JF (2002) Rapid diversification of the cotton genus (Gossypium: Malvaceae) revealed by analysis of sixteen nuclear and chloroplast genes. Am J Bot 89(4):707–725
Daniell H, Lee SB, Grevich J, Saski C, Quesada-Vargas T, Guda C, Tomkins J, Jansen RK (2006) Complete chloroplast genome sequences of Solanum bulbocastanum, Solanum lycopersicum and comparative analyses with other Solanaceae genomes. Theor Appl Genet 112(8):1503–1518
De Camargo GM, Porto-Neto LR, Kelly MJ, Bunch RJ, Mcwilliam SM, Tonhati H, Lehnert SA, Fortes MR, Moore SS (2015) Non-synonymous mutations mapped to chromosome X associated with andrological and growth traits in beef cattle. BMC Genom 16:384
Dong WL, Wang RN, Zhang NY, Fan WB, Fang MF, Li ZH (2018) Molecular evolution of chloroplast genomes of orchid species: insights into phylogenetic relationship and adaptive evolution. Int J Mol Sci 19(3):716
Fan WB, Wu Y, Yang J, Shahzad K, Li ZH (2018) Comparative chloroplast genomics of dipsacales species: insights into sequence variation, adaptive evolution, and phylogenetic relationships. Front Plant Sci 9:689
Flagel LE, Wendel JF (2010) Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol 186(1):184–193
Foster PG, Jermiin LS, Hickey DA (1997) Nucleotide composition bias affects amino acid content in proteins coded by animal mitochondria. J Mol Evol 44(3):282–288
Gogarten JP, Doolittle WF, Lawrence JG (2002) Prokaryotic evolution in light of gene transfer. Mol Biol Evol 19(12):2226–2238
Gray MW (1989) Origin and evolution of mitochondrial DNA. Annu Rev Cell Biol 5:25–50
Grover CE, Gallagher JP, Jareczek JJ, Page JT, Udall JA, Gore MA, Wendel JF (2015) Re-evaluating the phylogeny of allopolyploid Gossypium L. Mol Phylogenet Evol 92:45–52
Gruenstaeudl M, Nauheimer L, Borsch T (2017) Plastid genome structure and phylogenomics of Nymphaeales: conserved gene order and new insights into relationships. Plant Syst Evol 303(9):1251–1270
Hao J, Sun YQ (2015) Applied plant genomics and biotechnology. In: Poltronieri P (ed) Cotton genomics and biotechnology, 13th Woodhead Publishing, pp 213–227.
Hasenöhrl D, Fabbretti A, Londei P, Gualerzi CO, Bläsi U (2009) Translation initiation complex formation in the crenarchaeon Sulfolobus solfataricus. RNA 15(12):2288–2298
Hoang CV, Wessler HG, Local A, Turley RB, Benjamin RC, Chapman KD (1999) Identification and expression of cotton (Gossypium hirsutum L.) plastidial carbonic anhydrase. Plant Cell Physiol 40(12):1262–1270
Joy KW, Ireland RJ, Lea PJ (1983) Asparagine synthesis in pea leaves, and the occurrence of an asparagine synthetase inhibitor. Plant Physiol 73(1):165–168
Ke X, Cardon LR (2003) Efficient selective screening of haplotype tag SNPs. Bioinformatics 19(2):287–288
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9(4):299–306
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Lane N (2011) Plastids, genomes, and the probability of gene transfer. Genome Biol Evol 3:372–374
Larson SR, Doebley J (1994) Restriction site variation in the chloroplast genome of Tripsacum (Poaceae): phylogeny and rates of sequence evolution. Syst Bot 19(1):21–34
Lee S-B, Kaittanis C, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell H (2006) The complete chloroplast genome sequence of Gossypium hirsutum: organization and phylogenetic relationships to other angiosperms. BMC Genom 7:61
Lei BB, Li SS, Liu GZ, Chen ZW, Su AG, Li PB, Li ZH, Hua JP (2013) Evolution of mitochondrial gene content: loss of genes, tRNAs and introns between Gossypium harknessii and other plants. Plant Syst Evol 299(10):1889–1897
Liao BY, Scott NM, Zhang JZ (2006) Impacts of gene essentiality, expression pattern, and gene compactness on the evolutionary rate of mammalian proteins. Mol Biol Evol 23(11):2072–2080
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11):1451–1452
Liu GZ, Cao DD, Li SS, Su AG, Geng JN, Grover CE, Hu SN, Hua JP (2013) The complete mitochondrial genome of Gossypium hirsutum and evolutionary analysis of higher plant mitochondrial genomes. PLoS ONE 8(8):e69476
Logacheva MD, Samigullin TH, Dhingra A, Penin AA (2008) Comparative chloroplast genomics and phylogenetics of Fagopyrum esculentum ssp. Ancestrale—a wild ancestor of cultivated buckwheat. BMC Plant Biol 8:59
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25(5):955–964
Ma PF, Zhang YX, Guo ZH, Li DZ (2015) Evidence for horizontal transfer of mitochondrial DNA to the plastid genome in a bamboo genus. Sci Rep 5:11608
Martin W (2003) Gene transfer from organelles to the nucleus: frequent and in big chunks. Proc Natl Acad Sci USA 100(15):8612–8614
Martin W, Herrmann RG (1998) Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol 118:9–17
Matsuoka Y, Yamazaki Y, Ogihara Y, Tsunewaki K (2002) Whole chloroplast genome comparison of rice, maize, and wheat: implications for chloroplast gene diversification and phylogeny of cereals. Mol Biol Evol 19(12):2084–2091
Mcbain CJ, Mayer ML (1994) N-methyl-d-aspartic acid receptor structure and function. Physiol Rev 4(3):723–760
Meng ZL, Zaykin DV, Xu CF, Wagner M, Ehm MG (2003) Selection of genetic markers for association analyses, using linkage disequilibrium and haplotypes. Am J Hum Genet 73(1):115–130
Mes THM, Kuperus P, Kirschner J, Stepanek J, Oosterveld P, Storchova H, den Nijs JCM (2000) Hairpins involving both inverted and direct repeats are associated with homoplasious indels in non-coding chloroplast DNA of Taraxacum, (Lactuceae: Asteraceae). Genome 43(4):634–641
Miyata S, Nakazono M, Hirai A (1998) Transcription of plastid-derived tRNA genes in rice mitochondria. Curr Genet 34(3):216–220
Morton BR (1993) chloroplast DNA codon use: Evidence for selection at the psbA locus based on tRNA availability. J Mol Evol 37(3):273–280
Nachman MW, Crowell SL (2000) Estimate of the mutation rate per nucleotide in humans. Genetics 156(1):297–304
Percudani R, Ottonello S (1999) Selection at the wobble position of codons read by the same tRNA in Saccharomyces cerevisiae. Mol Biol Evol 16(12):1752–1762
Rota-Stabelli O, Lartillot N, Philippe H, Pisani D (2013) Serine codon-usage bias in deep phylogenomics: Pancrustacean relationships as a case study. Syst Biol 62(1):121–133
Saha S, Karaca M, Jenkins JN, Zipf AE, Reddy OUK, Kantety RV (2003) Simple sequence repeats as useful resources to study transcribed genes of cotton. Euphytica 130(3):355–364
Saini AK, Nanda JS, Lorsch JR, Hinnebusch AG (2010) Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNA(i)(Met) binding to the ribosome. Genes Dev 24(1):97–110
Saitou N, Ueda S (1994) Evolutionary rates of insertion and deletion in noncoding nucleotide sequences of primates. Mol Biol Evol 11(3):504–512
Sancho R, Cantalapiedra CP, López-Alvarez D, Gordon SP, Vogel JP, Catalán P, Contreras-Moreira B (2017) Comparative plastome genomics and phylogenomics of Brachypodium: flowering time signatures, introgression and recombination in recently diverged ecotypes. New Phytol 218(4):1631–1644
Saski C, Lee SB, Fjellheim S, Guda C, Jansen RK, Luo H, Tomkins J, Rognli OA, Daniell H, Clarke JL (2007) Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor Appl Genet 115(4):571–590
Sharp PM, Cowe E (1991) Synonymous codon usage in Saccharomyces cerevisiae. Yeast 7(7):657–678
Sharp PM, Li WH (1987) The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 15(3):1281–1295
Sloan DB, Alverson AJ, Wu M, Palmer JD, Taylor DR (2012) Recent acceleration of plastid sequence and structural evolution coincides with extreme mitochondrial divergence in the angiosperm genus silene. Genome Biol Evol 4(3):294–306
Smith DR (2011) Extending the limited transfer window hypothesis to inter-organelle DNA migration. Genome Biol Evol 3:743–748
Talat F, Wang KB (2015) Comparative bioinformatics analysis of the chloroplast genomes of a wild diploid Gossypium and two cultivated allotetraploid species. Iran J Biotechnol 13(3):47–56
Tang DT, Glazov EA, McWilliam SM, Barris WC, Dalrymple BP (2009) Analysis of the complement and molecular evolution of tRNA genes in cow. BMC Genom 10:188
Tang MY, Chen ZW, Grover CE, Wang YM, Li SS, Liu GZ, Ma ZY, Wendel JF, Hua JP (2015) Rapid evolutionary divergence of Gossypium barbadense and G. hirsutum mitochondrial genomes. BMC Genom 16:770
Treangen TJ, Rocha EPC (2011) Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet 7(1):e1001284
Tsai CT, Lin CH, Chang CY (2007) Analysis of codon usage bias and base compositional constraints in iridovirus genomes. Virus Res 126(1–2):196–206
Turmel M, Otis C, Lemieux C (2016) Mitochondrion-to-chloroplast dna transfers and intragenomic proliferation of chloroplast group ii introns in Gloeotilopsis green algae (Ulotrichales, Ulvophyceae). Genome Biol Evol 8(9):2789–2805
Wang YH, Huang CL, Jia JF (2009) Construction of multicistron expression vector by using plastid homologous sequences of sweet potato. Genom Appl Biol 28(4):659–667
Wang Y, Zhu FC, He LS, Danchin A (2018) Unique tRNA gene profile suggests paucity of nucleotide modifications in anticodons of a deep-sea symbiotic spiroplasma. Nucleic Acids Res 46(5):2197–2203
Wendel JF, Cronn RC (2003) Polyploidy and the evolutionary history of cotton. Adv Agron 78:139–186
Wolfe KH, Sharp PM (1988) Identification of functional open reading frames in chloroplast genomes. Gene 66(2):215–222
Wolfe KH, Li WH, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA 84(24):9054–9058
Wu Y, Liu F, Yang DG, Li W, Zhou XJ, Pei XY, Liu YG, He KL, Zhang WS, Ren ZY, Zhou KH, Ma XF, Li ZH (2018) Comparative chloroplast genomics of Gossypium species: insights into repeat sequence variations and phylogeny. Front Plant Sci 9:376
Xia X, Luan LL, Qin G, Yu LF, Wang ZW, Dong WC, Song Y, Qiao Y, Zhang XS, Sang YL, Yang L (2017) Genome-wide analysis of SSR and ILP markers in trees: diversity profiling, alternate distribution, and applications in duplication. Sci Rep 7(1):17902
Xu Q, Xiong GJ, Li PB, He F, Huang Y, Wang KB, Li ZH, Hua JP (2012) Analysis of complete nucleotide sequences of 12 Gossypium chloroplast genomes: origin and evolution of allotetraploids. PLoS ONE 7(8):e37128
Yang L, Gaut BS (2011) Factors that contribute to variation in evolutionary rate among Arabidopsis genes. Mol Biol Evol 28(8):2359–2369
Zhang G, Hubalewska M, Ignatova Z (2009) Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol 16(3):274–280
Zhang MY, Fritsch PW, Ma PF, Wang H, Lu L, Li DZ (2017) Plastid phylogenomics and adaptive evolution of Gaultheria series Trichophyllae (Ericaceae), a clade from sky islands of the Himalaya-Hengduan mountains. Mol Phylogenet Evol 110:7–18
Zhou M, Long W, Li X (2008) Patterns of synonymous codon usage bias in chloroplast genomes of seed plants. For Stud China 11(4):235–242
Funding
This research was funded by grants from National Key R & D Program for Crop Breeding (2016YFD0100306), National Natural Science Foundation of China (No. 31401431), and the Shaanxi Science and Technology Innovation Team (2019TD‐012), the Public health specialty in the Department of traditional Chinese Medicine (Grant nos. 2017-66 and 2018-43) and the Open Foundation of Key Laboratory of Resource Biology and Biotechnology in Western China (Ministry of Education) (Grant nos. ZSK2017007 and ZSK2019008).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Amit Dhingra .
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
299_2020_2529_MOESM1_ESM.tif
Supplementary file1 Secondary structures inferred for the tRNAs present in the Gossypium chloroplast genomes (TIF 1960 kb)
Rights and permissions
About this article
Cite this article
Zhang, TT., Liu, H., Gao, QY. et al. Gene transfer and nucleotide sequence evolution by Gossypium cytoplasmic genomes indicates novel evolutionary characteristics. Plant Cell Rep 39, 765–777 (2020). https://doi.org/10.1007/s00299-020-02529-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-020-02529-9