Abstract
Key message
The chloroplast-localized protein CSAP is an ABA-responsive factor and positively regulates dark-induced senescence. This phenomenon is controlled by SAUL1 in Arabidopsis.
Abstract
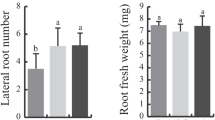
We report here that CSAP (Chloroplast-localized Senescence-Associated Protein, AT5G39520) functions as a positive regulator of senescence and is controlled by SAUL1 (Senescence Associated E3 Ubiquitin Ligase 1) in Arabidopsis. CSAP transcript level was gradually increased when senescence was progressed. Under dark conditions, the csap mutant showed delayed leaf senescence and reduced chlorophyll breakdown, but overexpression of CSAP accelerated leaf senescence and expressions of chlorophyll catabolic genes were up-regulated compared to the wild-type (WT). NCED3 and AAO3, which are involved in ABA biosynthesis, also showed higher expression in the overexpression lines than the WT. It is known that the CSAP transcript is increased in the saul1 mutant that shows precocious senescence. In our experiments, we confirmed that CSAP interacts with SAUL1 by the yeast two-hybrid and pull-down assays. In addition, we found that SAUL1 decreases the stability of CSAP in the presence of ABA. Taken together, we suggest that CSAP accelerates leaf senescence in the dark and this process is controlled by SAUL1.





Similar content being viewed by others
Abbreviations
- CSAP:
-
Chloroplast-localized senescence-associated protein
- SAUL1:
-
Senescence-associated E3 ubiquitin ligase 1
- ABA:
-
Abscisic acid
- WT:
-
Wild type
- OX:
-
Overexpressed
- KO:
-
Knock-out
- GUS:
-
β-glucuronidase
- qRT-PCR:
-
Quantitative reverse transcription-polymerase chain reaction
References
Arnon D (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol 24:1–15
Balazadeh S, Siddiqui H, Allu AD et al (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62:250–264
Balazadeh S, Kwasniewski M, Caldana C et al (2011) ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant 4:346–360
Carrión CA, Martínez DE, Costa ML, Guiamet JJ (2014) Senescence-associated vacuoles, a specific lytic compartment for degradation of chloroplast proteins? Plants 3:498–512
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Drechsel G, Bergler J, Wippel K et al (2011) C-terminal armadillo repeats are essential and sufficient for association of the plant U-box armadillo E3 ubiquitin ligase SAUL1 with the plasma membrane. J Exp Bot 62:775–785
Gao S, Gao J, Zhu X et al (2016) ABF2, ABF3, and ABF4 promote ABA-mediated chlorophyll degradation and leaf senescence by transcriptional activation of chlorophyll catabolic genes and senescence-associated genes in Arabidopsis. Mol Plant 9:1272–1285
Garapati P, Xue G-P, Munné-Bosch S, Balazadeh S (2015) Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol 168:1122–1139
Guo Y, Gan S-S (2012) Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant, Cell Environ 35:655–664
He Y, Fukushige H, Hildebrand DF, Gan S (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128:876–884
Hensel LL, Grbić V, Baumgarten DA, Bleecker AB (1993) Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell 5:553–564
Hörtensteiner S, Feller U (2002) Nitrogen metabolism and remobilization during senescence. J Exp Bot 53:927–937
Hörtensteiner S, Kräutler B (2011) Chlorophyll breakdown in higher plants. BBA 1807:977–988
Jensen MK, Lindemose S, de Masi F et al (2013) ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio 3:321–327
Jibran R, Hunter DA, Dijkwel PP (2013) Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol Biol 82:547–561
Jin JB, Kim YA, Kim SJ et al (2001) A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13:1511–1525
Khan M, Rozhon W, Poppenberger B (2014) The role of hormones in the aging of plants-a mini-review. Gerontology 60:49–55
Kim J, Kim JH, Lyu JI et al (2018) New insights into the regulation of leaf senescence in Arabidopsis. J Exp Bot 69:787–799
Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204:383–396
Lee IH, Lee IC, Kim J et al (2016) NORE1/SAUL1 integrates temperature-dependent defense programs involving SGT1b and PAD4 pathways and leaf senescence in Arabidopsis. Physiol Plant 158:180–199
Liebsch D, Keech O (2016) Dark-induced leaf senescence: new insights into a complex light-dependent regulatory pathway. New Phytol 212:563–570
Lim PO, Woo HR, Nam HG (2003) Molecular genetics of leaf senescence in Arabidopsis. Trends Plant Sci 8:272–278
Liu X, Li Z, Jiang Z et al (2011) LSD: a leaf senescence database. Nuclc Acids Res 39:D1103–D1107
Liu T, Longhurst AD, Talavera-Rauh F et al (2016) The Arabidopsis transcription factor ABIG1 relays ABA signaled growth inhibition and drought induced senescence. eLife 5:e13768
Lohman KN, Gan S, John MC, Amasino RM (1994) Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol Plant 92:322–328
Morris K, Mackerness SAH, Page T et al (2000) Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J 23:677–685
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Oh SA, Park J-H, Lee IG et al (1997) Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J 12:527–535
Raab S, Drechsel G, Zarepour M et al (2009) Identification of a novel E3 ubiquitin ligase that is required for suppression of premature senescence in Arabidopsis. Plant J 59:39–51
Sakuraba Y, Kim D, Paek N-C (2018) Salt treatments and induction of senescence. Plant Senescence 1744:141–149
Salt JN, Yoshioka K, Moeder W, Goring DR (2011) Altered germination and subcellular localization patterns for PUB44/SAUL1 in response to stress and phytohormone treatments. PLoS One 6:e21321
Streit L, Feller U (1983) Changing activities and different resistance to proteolytic activity of two forms of glutamine synthetase in wheat leaves during senescence. Physiol Veg 21:103–108
Thoenen M, Feller U (1998) Degradation of glutamine synthetase in intact chloroplasts isolated from pea (Pisum sativum) leaves. Aust J Plant Physiol 25:279–286
Verdaguer B, de Kochko A, Fux CI et al (1998) Functional organization of the cassava vein mosaic virus (CsVMV) promoter. Plant Mol Biol 37:1055–1067
Vogelmann K, Drechsel G, Bergler J et al (2012) Early senescence and cell death in Arabidopsis saul1 mutants involves the PAD4-dependent salicylic acid pathway. Plant Physiol 159:1477–1487
Wang S, Blumwald E (2014) Stress-induced chloroplast degradation in Arabidopsis is regulated via a process independent of autophagy and senescence-associated vacuoles. Plant Cell 26:4875–4888
Wu Y, Deng Z, Lai J (2009) Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res 19:1279–1290
Yang J, Worley E, Udvardi M (2014) A NAP-AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell 26:4862–4874
Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2:1565–1572
Zhao Y, Chan Z, Gao J et al (2016) ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci USA 113:1949–1954
Acknowledgements
This work was supported by a grant (PJ01367001) from the Next-Generation BioGreen 21 Program funded by the Rural Development Administration, Republic of Korea. This work was also partially supported by Korea University.
Author information
Authors and Affiliations
Contributions
WMS and JSS conceived the study. WMS, SYK and JSS designed experiments. WMS, SYK and SH performed experiments. WMS, SYK and JSS wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Youn-Il Park.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
So, W.M., Kim, S.Y., Hyoung, S. et al. The novel protein CSAP accelerates leaf senescence and is negatively regulated by SAUL1 in the dark. Plant Cell Rep 39, 325–334 (2020). https://doi.org/10.1007/s00299-019-02493-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-019-02493-z