Abstract
Key message
GbWRKY1 can function as a negative regulator of ABA signaling via JAZ1 and ABI1, with effects on salt and drought tolerance.
Abstract
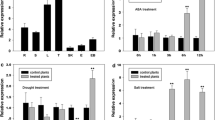
WRKY transcription factors play important roles in plant development and stress responses. GbWRKY1 was initially identified as a defense-related gene in cotton and negatively regulates the response to fungal pathogens by activating the expression of JAZ1. Here, we characterized the role of GbWRKY1, an orthologue of the Arabidopsis gene AtWRKY75, in abiotic stress (salt and drought) and established novel connection between JAZ1 and ABA signaling in Arabidopsis. GbWRKY1 is nucleus localized and its expression is significantly induced by treatment with ABA and osmotic stresses NaCl and PEG. Increased levels of expression of GbWRKY1 in transgenic Arabidopsis enhance sensitivity to salt and drought as revealed by seed germination tests and soil stress experiments. Similarly, GbWRKY1 overexpression cotton plants also display increased sensitivity to PEG treatment and drought. Expression analysis shows that the induction of two ABA responsive genes, RAB18 and RD29A by NaCl, mannitol, and ABA treatment is significantly impaired in GbWRKY1 overexpression Arabidopsis lines. GbWRKY1 overexpression Arabidopsis displays a strong ABA-insensitive phenotype at both germination and early stages of seedling development. Further genetic evidence suggested that the ABA-insensitive phenotype of GbWRKY1 overexpression Arabidopsis was dependent on JAZ1, and overexpression of JAZ1 also displayed an ABA-insensitive phenotype. In addition, yeast two hybrid and bimolecular fluorescence complementation assays showed that JAZ1 directly interacts with ABI1, a key negative regulator of ABA signaling. We, therefore, demonstrate that GbWRKY1 acts as a negative regulator of ABA signaling, through an interaction network involving JAZ1 and ABI1, to regulate salt and drought tolerance.








Similar content being viewed by others
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Aleman F, Yazaki J, Lee M, Takahashi Y, Kim AY, Li ZX et al (2016) An ABA-increased interaction of the PYL6 ABA receptor with MYC2 transcription factor: a putative link of ABA and JA signaling. Sci Rep 6:28941
Bartels D, Sunkar R (2005) Drought and salt tolerance in plants. Crit Rev Plant Sci 24:23–58
Chen X, Yan D, Sun L, Zeng R, Yang L (2016) Cloning and expression analysis of HbWRKY75 gene in leaf from Hevea brasiliensis. Plant Physiol J 52:250–258
Dave A, Hernandez ML, He ZS, Andriotis VME, Vaistij FE, Larson TR, Graham IA (2011) 12-Oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell 23:583–599
de Ollas C, Arbona V, Gomez-Cadenas A (2015) Jasmonoyl isoleucine accumulation is needed for abscisic acid build-up in roots of Arabidopsis under water stress conditions. Plant Cell Environ 38:2157–2170
Devaiah BN, Karthikeyan AS, Raghothama KG (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143:1789–1801
Ellis C, Turner JG (2002) A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215:549–556
Fernandez-Arbaizar A, Regalado JJ, Lorenzo O (2012) Isolation and characterization of novel mutant loci suppressing the ABA hypersensitivity of the Arabidopsis Coronatine insensitive 1–16 (coi1-16) mutant during germination and seedling growth. Plant Cell Physiol 53:53–63
Golldack D, Luking I, Yang O (2011) Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep 30:1383–1391
Guo PR, Li ZH, Huang PX, Li BS, Fang S, Chu JF, Guo HW (2017) A tripartite amplification loop involving the transcription factor WRKY75, Salicylic Acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 29:2854–2870
Hecker A, Wallmeroth N, Peter S, Blatt MR, Harter K, Grefen C (2015) Binary 2in1 vectors improve in planta (co)localization and dynamic protein interaction studies. Plant Physiol 168:776–787
Hossain MA, Henriquez-Valencia C, Gomez-Paez M, Medina J, Orellana A, Vicente-Carbajosa J, Zouhar J (2016) Identification of novel components of the unfolded protein response in Arabidopsis. Front Plant Sci 7:650
Jiang JJ, Ma SH, Ye NH, Jiang M, Cao JS, Zhang JH (2017) WRKY transcription factors in plant responses to stresses. J Integr Plant Biol 59:86–101
Ju L, Jing Y, Shi P, Liu J, Chen J, Yan J et al (2019) JAZ proteins modulate seed germination through interaction with ABI5 in bread wheat and Arabidopsis. New Phytol 223:246–260
Kazan K, Manners JM (2012) JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci 17:22–31
Lackman P, Gonzalez-Guzman M, Tilleman S, Carqueijeiro I, Perez AC, Moses T et al (2011) Jasmonate signaling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in Arabidopsis and tobacco. Proc Natl Acad Sci USA 108:5891–5896
Li C, He X, Luo XY, Xu L, Liu LL, Min L et al (2014) Cotton WRKY1 Mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol 166:2179–2194
Liu H, Ding YD, Zhou YQ, Jin WQ, Xie KB, Chen LL (2017) CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants. Mol Plant 10:530–532
Liu Q, Wang C, Jiao XZ, Zhang HW, Song LL, Li YX et al (2019) Hi-TOM: a platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci China Life Sci 62:1–7
Long L, Yang WW, Liao P, Guo YW, Kumar A, Gao W (2019) Transcriptome analysis reveals differentially expressed ERF transcription factors associated with salt response in cotton. Plant Sci 281:72–81
Luo X, Bai X, Sun XL, Zhu D, Liu BH, Ji W et al (2013) Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J Exp Bot 64:2155–2169
Meyer K, Leube MP, Grill E (1994) A Protein Phosphatase 2C involved in ABA signal-transduction in Arabidopsis thaliana. Science 264:1452–1455
Nakashima K, Yamaguchi-Shinozaki K (2013) ABA signaling in stress-response and seed development. Plant Cell Rep 32:959–970
Ou B, Yin KQ, Liu SN, Yang Y, Gu T, Hui JMW et al (2011) A high-throughput screening system for arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Mol Plant 4:546–555
Parihar P, Singh S, Singh R, Singh VP, Prasad SM (2015) Effect of salinity stress on plants and its tolerance strategies: a review. Environ Sci Pollut R 22:4056–4075
Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y et al (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL Family of START proteins. Science 324:1068–1071
Raghavendra AS, Gonugunta VK, Christmann A, Grill E (2010) ABA perception and signalling. Trends Plant Sci 15:395–401
Ranjan A, Sawant S (2015) Genome-wide transcriptomic comparison of cotton (Gossypium herbaceum) leaf and root under drought stress. 3 Biotech 5:585–596
Ranjan A, Pandey N, Lakhwani D, Dubey NK, Pathre UV, Sawant SV (2012) Comparative transcriptomic analysis of roots of contrasting Gossypium herbaceum genotypes revealing adaptation to drought. BMC Genom 13:680
Rushton PJ, Somssich IE, Ringler P, Shen QXJ (2010) WRKY transcription factors. Trends Plant Sci 15:247–258
Sah SK, Reddy KR, Li JX (2016) Abscisic acid and abiotic stress tolerance in crop plants. Front Plant Sci 7:571
Shao HB, Wang HY, Tang XL (2015) NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front Plant Sci 6:902
Shi WN, Hao LL, Li J, Liu DD, Guo XQ, Li H (2014) The Gossypium hirsutum WRKY gene GhWRKY39-1 promotes pathogen infection defense responses and mediates salt stress tolerance in transgenic Nicotiana benthamiana. Plant Cell Rep 33:483–498
Singh D, Laxmi A (2015) Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front Plant Sci 6:895
Song Y, Chen LG, Zhang LP, Yu DQ (2010) Overexpression of OsWRKY72 gene interferes in the abscisic acid signal and auxin transport pathway of Arabidopsis. J Biosciences 35:459–471
Ullah A, Sun H, Yang XY, Zhang XL (2017) Drought coping strategies in cotton: increased crop per drop. Plant Biotechnol J 15:271–284
Verma V, Ravindran P, Kumar PP (2016) Plant hormone-mediated regulation of stress responses. BMC Plant Biol 16:86
Wan Y, Mao M, Wan D, Liu J, Wang G, Li G, Wang R (2018) Caragana intermedia WRKY75 Altered Arabidopsis thaliana tolerance to salt stress and ABA. Acta Bot Boreali-Occidential Sinica 38:17–25
Wang PC, Zhang J, Sun L, Ma YZ, Xu J, Liang SJ et al (2018) High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol J 16:137–150
Xu L, Jin L, Long L, Liu LL, He X, Gao W et al (2012) Overexpression of GbWRKY1 positively regulates the Pi starvation response by alteration of auxin sensitivity in Arabidopsis. Plant Cell Rep 31:2177–2188
Zhang SC, Li C, Wang R, Chen YX, Shu S, Huang RH et al (2017) The Arabidopsis mitochondrial protease FtSH4 is involved in leaf senescence via regulation of WRKY-dependent salicylic acid accumulation and signaling. Plant Physiol 173:2294–2307
Zhang LL, Cheng J, Sun XM, Zhao TT, Li MJ, Wang QF et al (2018a) Overexpression of VaWRKY14 increases drought tolerance in Arabidopsis by modulating the expression of stress-related genes. Plant Cell Re 37:1159–1172
Zhang LP, Chen LG, Yu DQ (2018b) Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol 176:790–803
Acknowledgments
We thank Prof. Keith Lindsey (Durham University) for critical suggestions on manuscript revision. We are grateful to Huazhi Song and De Zhu (National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University) for confocal imaging, and Pengcheng Wang (National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University) for providing the CRISPR vector. We thank Prof. Sheng Luan and Dr. Congcong Hou (University of California, Berkeley) for providing pDONR221-P1P4, pDONR221-P3P2, pBIFCt-2in1-NN, and pFRETgc-2in1-NN plasmids.
Funding
This work was financially supported by the National Natural Science Foundation of China (31471541) and the National Key Project of Research and Development (2018YFD0100403).
Author information
Authors and Affiliations
Contributions
XZ, LZ, and XL conceived and designed research. XL conducted experiments and wrote the manuscript. CL generated GbWRKY1 transgenic materials. XH gave constructive suggestions on stress experiments. LZ revised the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors have no conflicts of interest to declare.
Additional information
Communicated by Chun-Hai Dong.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Luo, X., Li, C., He, X. et al. ABA signaling is negatively regulated by GbWRKY1 through JAZ1 and ABI1 to affect salt and drought tolerance. Plant Cell Rep 39, 181–194 (2020). https://doi.org/10.1007/s00299-019-02480-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-019-02480-4