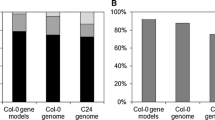
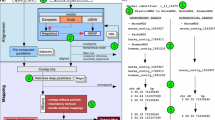
Abstract
Key message
Polymorphic probes identified via a sequence-based approach are suitable to infer the genotypes of recombinant inbred lines from hybridisation intensities of GeneChip ® transcript profiling experiments.
Abstract
The sequences of the probes of the ATH1 GeneChip® exactly match transcript sequences of the Arabidopsis thaliana reference genome Col-0, whereas nucleotide differences and/or insertions/deletions may be observed for transcripts of other A. thaliana accessions. Individual probes of the GeneChip® that show sequence polymorphisms between different A. thaliana accessions may serve as single-feature polymorphism (SFP) markers, provided that the sequence changes cause differences in hybridisation intensity for the accessions of interest. A sequence-based approach identified features on the high-density oligonucleotide array that showed sequence polymorphisms between A. thaliana accessions Col-0 and C24. Hybridisation intensities of polymorphic probes were extracted from genome-wide transcript profiles of Col-0/C24 and C24/Col-0 recombinant inbred lines and assessed after standardisation via sliding window analyses to identify SFP markers. The genotypes of the recombinant inbred lines were determined with the SFP markers and the resulting data were integrated with information, which had been established previously with single nucleotide polymorphism and insertion/deletion markers, to enrich the linkage map of the Col-0/C24 and C24/Col-0 recombinant inbred populations. Congruence between the molecular marker map and the sequence maps of the A. thaliana Col-0 chromosomes proved the reliability of the genotype information which was deduced from the transcript profiles of the Col-0/C24 and C24/Col-0 recombinant inbred lines.



Similar content being viewed by others
Abbreviations
- DAF:
-
Days after flower opening
- eQTL:
-
Expression quantitative trait loci
- GS:
-
Gap size
- GEO:
-
Gene Expression Omnibus
- IM:
-
Invariant match
- Indel:
-
Insertion/deletion
- PM:
-
Perfect match
- RA:
-
Ratio
- RIL:
-
Recombinant inbred line
- SFP:
-
Single-feature polymorphism
- SNP:
-
Single nucleotide polymorphism
- VM:
-
Variant match
- WS:
-
Window size
References
Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, Rudnev D, Lash AE, Fujibuchi W, Edgar R (2005) NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res 33(Database issue):D562–D566
Bergelson J, Buckler ES, Ecker JR, Nordborg M, Weigel D (2016) A proposal regarding best practices for validating the identity of genetic stocks and the effects of genetic variants. Plant Cell 28:606–609
Borevitz JO, Liang D, Plouffe D, Chang HS, Zhu T, Weigel D, Berry CC, Winzeler E, Chory J (2003) Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res 13:513–523
Boudichevskaia A, Cao HX, Schmidt R (2017) Tailoring high-density oligonucleotide arrays for transcript profiling of different Arabidopsis thaliana accessions using a sequence-based approach. Plant Cell Rep 36:1323–1332
Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, Wang X, Ott F, Müller J, Alonso-Blanco C, Borgwardt K, Schmid KJ, Weigel D (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43:956–963
Cui X, Xu J, Asghar R, Condamine P, Svensson JT, Wanamaker S, Stein N, Roose M, Close TJ (2005) Detecting single-feature polymorphisms using oligonucleotide arrays and robustified projection pursuit. Bioinformatics 21:3852–38588
Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, Kahles A, Bohnert R, Jean G, Derwent P, Kersey P, Belfield EJ, Harberd NP, Kemen E, Toomajian C, Kover PX, Clark RM, Rätsch G, Mott R (2011) Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477:419–423
Gresham D, Ruderfer DM, Pratt SC, Schacherer J, Dunham MJ, Botstein D, Kruglyak L (2006) Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science 311:1932–1936
Gupta PK, Rustgi S, Mir RR (2008) Array-based high-throughput DNA markers for crop improvement. Heredity 101:5–18
He S, Zhao Y, Mette MF, Bothe R, Ebmeyer E, Sharbel TF, Reif JC, Jiang Y (2015) Prospects and limits of marker imputation in quantitative genetic studies in European elite wheat (Triticum aestivum L.). BMC Genom 16:168
He F, Yoo S, Wang D, Kumari S, Gerstein M, Ware D, Maslov S (2016) Large-scale atlas of microarray data reveals the distinct expression landscape of different tissues in Arabidopsis. Plant J 86:472–480
Howie BN, Donnelly P, Marchini J (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5(6):e1000529
Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31:e15
Jansen RC, Nap JP (2001) Genetical genomics: the added value from segregation. Trends Genet 17:388–391
Knoch D, Riewe D, Meyer R, Boudichevskaia A, Schmidt R, Altmann T (2017) Genetic dissection of metabolite variation in Arabidopsis seeds: evidence for mQTL hotspots and a master regulatory locus. J Exp Bot 68:1655–1667
Kumar R, Qiu J, Joshi T, Valliyodan B, Xu D, Nguyen HT (2007) Single feature polymorphism discovery in rice. PLoS One 2:e284
Langmead B, Trapnell C, Pop M, Salzberg S (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Lisec J, Meyer RC, Steinfath M, Redestig H, Becher M, Witucka-Wall H, Fiehn O, Törjék O, Selbig J, Altmann T, Willmitzer L (2008) Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J 53:960–972
Lisec J, Steinfath M, Meyer RC, Selbig J, Melchinger AE, Willmitzer L, Altmann T (2009) Identification of heterotic metabolite QTL in Arabidopsis thaliana RIL and IL populations. Plant J 59:777–788
Meyer RC, Kusterer B, Lisec J, Steinfath M, Becher M, Scharr H, Melchinger AE, Selbig J, Schurr U, Willmitzer L, Altmann T (2010) QTL analysis of early stage heterosis for biomass in Arabidopsis. Theor Appl Genet 120:227–237
Rostoks N, Borevitz JO, Hedley PE, Russell J, Mudie S, Morris J, Cardle L, Marshall DF, Waugh R (2005) Single-feature polymorphism discovery in the barley transcriptome. Genome Biol 6:R54
Schneeberger K, Ossowski S, Ott F, Klein JD, Wang X, Lanz C, Smith LM, Cao J, Fitz J, Warthmann N, Henz SR, Huson DH, Weigel D (2011) Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc Natl Acad Sci USA 108:10249–10254
Singer T, Fan Y, Chang HS, Zhu T, Hazen SP, Briggs SP (2006) A high-resolution map of Arabidopsis recombinant inbred lines by whole-genome exon array hybridization. PLoS Genet 2:e144
Stange M, Utz HF, Schrag TA, Melchinger AE, Würschum T (2013) High-density genotyping: an overkill for QTL mapping? Lessons learned from a case study in maize and simulations. Theor Appl Genet 126:2563–2574
The 1001 Genomes Consortium (2016) 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166:481–491
The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Todesco M, Balasubramanian S, Cao J, Ott F, Sureshkumar S, Schneeberger K, Meyer RC, Altmann T, Weigel D (2012) Natural variation in biogenesis efficiency of individual Arabidopsis thaliana microRNAs. Curr Biol 22:166–170
Törjék O, Witucka-Wall H, Meyer RC, von Korff M, Kusterer B, Rautengarten C, Altmann T (2006) Segregation distortion in C24/Col-0 and Col-0/C24 recombinant inbred line populations is due to reduced fertility caused by epistatic interactions of two loci. Theor Appl Genet 113:1551–1561
van Os H, Stam P, Visser RG, van Eck HJ (2005) SMOOTH: a statistical method for successful removal of genotyping errors from high-density genetic linkage data. Theor Appl Genet 112:187–194
West MA, van Leeuwen H, Kozik A, Kliebenstein DJ, Doerge RW, St Clair DA, Michelmore RW (2006) High-density haplotyping with microarray-based expression and single feature polymorphism markers in Arabidopsis. Genome Res 16:787–795
Winzeler EA, Richards DR, Conway AR, Goldstein AL, Kalman S, McCullough MJ, McCusker JH, Stevens DA, Wodicka L, Lockhart DJ, Davis RW (1998) Direct allelic variation scanning of the yeast genome. Science 281:1194–1197
Wu Y, Bhat PR, Close TJ, Lonardi S (2008) Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet 4(10):e1000212
Xie W, Chen Y, Zhou G, Wang L, Zhang C, Zhang J, Xiao J, Zhu T, Zhang Q (2009) Single feature polymorphisms between two rice cultivars detected using a median polish method. Theor Appl Genet 119:151–164
Xu WW, Cho S, Yang SS, Bolon YT, Bilgic H, Jia H, Xiong Y, Muehlbauer GJ (2007) Single-feature polymorphism discovery by computing probe affinity shape powers. BMC Genet 10:48
Zhu T, Salmeron J (2007) High-definition genome profiling for genetic marker discovery. Trends Plant Sci 12:196–202
Acknowledgements
Aspects of this study were funded through BMBF grant GABI-OIL (0315053G).The authors thank Dmitri Pescianschi for support during the custom C++ script development and Jahnavi Koppolu for her contribution to the analysis of plant phenotypes. Kristin Langanke and Angelika Flieger are acknowledged for skilful technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Dr. Jim Register.
Electronic supplementary material
Below is the link to the electronic supplementary material.
299_2017_2200_MOESM2_ESM.xlsx
Supplementary file 2: RA values of recombinant inbred lines and parental lines Col-0 and C24. For the parental lines values for three replicates each are given, for the RILs a single replicate each was analysed. (XLSX 29898 kb)
299_2017_2200_MOESM4_ESM.xlsx
Supplementary file 4: Alignment of ATH1 probe sequences to the Col-0 and C24 genomes that provided the basis for SFP marker identification. Column “Best hit note” summarises the results of sequence alignments with Bowtie, only alignments containing up to three mismatches but no Indels are listed for each probe. IM, invariant match denotes a probe that shows unique perfect matches when compared to the Col-0 and C24 genomes; PM, unique perfect match; MisM, unique match containing between one and three mismatches but no Indel(s); NoM, no match or matches that contain Indels and/or more than three mismatches; VM, variant match refers to probes that reveals a unique perfect match when compared to the Col-0 genome whereas the C24 genome shows at least one mismatch or no alignment at all. For the VM probes the positions and identity of SNPs in the oligonucleotide sequences are indicated in the column “C24 SNPs”. (XLSX 6445 kb)
299_2017_2200_MOESM5_ESM.xlsx
Supplementary file 5: Graphical genotypes of recombinant inbred lines. Thresholds determined with different sliding window parameters were used to deduce the RIL genotypes for the different SFP markers. WS 5/GS 2* and WS 3/GS 1* refer to consensus genotype scores that were based on eight different parameter combinations of the sliding window analysis (WS 2/GS 2, WS 3/GS 1, WS 3/GP 2, WS 4/GS 1, WS 4/GS 2, WS 5/GS 0, WS 5/GS 1, WS 5/GS 2). SFP markers were ordered according to their position on the chromosome sequence maps of the reference accession Col-0 with the exception of the 23 unreliable SFP markers which are shown at the end of the compilation. Col-0 and C24 genotypes are denoted “a” and “b”, respectively. Putative double-recombination events that were identified by a single SFP marker are shown in italics and are marked with the postfix “-DR”. “NDR” marks those scores for which genotypes were not assigned. (XLSX 11659 kb)
299_2017_2200_MOESM7_ESM.xlsx
Supplementary file 7: Molecular marker map of recombinant inbred lines. The cumulative genetic distances are given for all markers which were used for the bin map in cM. For each marker it is indicated whether it represents an SNP, Indel or SFP marker. SFP marker names correspond to the probe set they were derived from (XLSX 26 kb)
Rights and permissions
About this article
Cite this article
Schmidt, R., Boudichevskaia, A., Cao, H.X. et al. Extracting genotype information of Arabidopsis thaliana recombinant inbred lines from transcript profiles established with high-density oligonucleotide arrays. Plant Cell Rep 36, 1871–1881 (2017). https://doi.org/10.1007/s00299-017-2200-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-017-2200-6