Abstract
Key message
Resistant and susceptible lines in Brassica rapa have different immune responses against Fusarium oxysporum inoculation.
Abstract
Fusarium yellows caused by Fusarium oxysporum f. sp. conglutinans (Foc) is an important disease of Brassicaceae; however, the mechanism of how host plants respond to Foc is still unknown. By comparing with and without Foc inoculation in both resistant and susceptible lines of Chinese cabbage (Brassica rapa var. pekinensis), we identified differentially expressed genes (DEGs) between the bulked inoculated (6, 12, 24, and 72 h after inoculation (HAI)) and non-inoculated samples. Most of the DEGs were up-regulated by Foc inoculation. Quantitative real-time RT-PCR showed that most up-regulated genes increased their expression levels from 24 HAI. An independent transcriptome analysis at 24 and 72 HAI was performed in resistant and susceptible lines. GO analysis using up-regulated genes at 24 HAI indicated that Foc inoculation activated systemic acquired resistance (SAR) in resistant lines and tryptophan biosynthetic process and responses to chitin and ethylene in susceptible lines. By contrast, GO analysis using up-regulated genes at 72 HAI showed the overrepresentation of some categories for the defense response in susceptible lines but not in the resistant lines. We also compared DEGs between B. rapa and Arabidopsis thaliana after F. oxysporum inoculation at the same time point, and identified genes related to defense response that were up-regulated in the resistant lines of Chinese cabbage and A. thaliana. Particular genes that changed expression levels overlapped between the two species, suggesting that they are candidates for genes involved in the resistance mechanisms against F. oxysporum.





Similar content being viewed by others
References
Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415:977–983
Beckers GJ, Spoel SH (2006) Fine-tuning plant defence signalling: salicylate versus jasmonate. Plant Biol 8:1–10
Berrocal-Lobo M, Molina A (2004) Ethylene response factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum. Mol Plant Microbe Interact 17:763–770
Beckman CH (1987) The nature of wilt diseases of plants. American Phytopathological Society, St. Paul
Bigeard J, Colcombet J, Hirt H (2015) Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant 8:521–539
Charoenporn C, Kanokmedhakul S, Lin FC, Poeaim S, Soytong K (2010) Evaluation of bio-agent formulations to control Fusarium wilt of tomato. Afr J Biotechnol 9:5836–5844
Chen Z, Agnew JL, Cohen JD, He P, Shan L, Sheen J, Kunkel BN (2007) Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci USA 104:20131–20136
Cheng F, Sun R, Hou X, Zheng H, Zhang F, Zhang Y, Liu B, Liang J, Zhuang M, Liu Y, Liu D, Wang X, Li P, Liu Y, Lin K, Bucher J, Zhang N, Wang Y, Wang H, Deng J, Liao Y, Wei K, Zhang X, Fu L, Hu Y, Liu J, Cai C, Zhang S, Zhang S, Li F, Zhang H, Zhang J, Guo N, Liu Z, Liu J, Sun C, Ma Y, Zhang H, Cui Y, Freeling MR, Borm T, Bonnema G, Wu J, Wang X (2016) Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat Genet 48:1218–1224
Daly P, Tomkins B (1995) Production and postharvest handling of Chinese cabbage (Brassica rapa var. pekinensis). RIRDC 97:41
Dangl JL, Jones JDG (2001) Plant pathogens and integrated defence responses to infection. Nature 411:826–833
Diener AC, Ausubel FM (2005) RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171:305–321
Dong J, Chen C, Chen Z (2003) Expression profiles of Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol 51:21–37
Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38:W64–W70
Enya J, Togawa M, Takeuchi T, Yoshida S, Tsushima S, Arie T, Sakai T (2008) Biological and phylogenetic characterization of Fusarium oxysporum complex, which causes yellows on Brassica spp., and proposal of F. oxysporum f. sp. rapae, a novel forma specialis pathogenic on B. rapa in Japan. Phytopathology 98:475–483
Fujimoto R, Sasaki T, Nishio T (2006) Characterization of DNA methyltransferase genes in Brassica rapa. Genes Genet Syst 81:235–242
Fujimoto R, Nishio T (2007) Self-incompatibility. Adv Bot Res 45:139–154
Gao QM, Venugopal S, Navarre D, Kachroo A (2011) Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol 155:464–476
Garcion C, Lohmann A, Lamodière E, Catinot J, Buchala A, Doermann P, Métraux JP (2008) Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol 147:1279–1287
Glazebrook J (2001) Genes controlling expression of defense responses in Arabidopsis–2001 status. Curr Opin Plant Biol 4:301–308
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227
Hu Y, Dong Q, Yu D (2012) Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci 185–186:288–297
Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323–329
Joshi RK, Nayak S (2011) Functional characterization and signal transduction ability of nucleotide-binding site-leucine-rich repeat resistance genes in plants. Genet Mol Res 10:2637–2652
Kawamura K, Kawanabe T, Shimizu M, Okazaki K, Kaji M, Dennis ES, Osabe K, Fujimoto R (2015) Genetic characterization of inbred lines of Chinese cabbage by DNA markers; towards the application of DNA markers to breeding of F1 hybrid cultivars. Data Brief 6:229–237
Kidd BN, Kadoo NY, Dombrecht B, Tekeoğlu M, Gardiner DM, Thatcher LF, Aitken EAB, Schenk PM, Manners JM, Kazan K (2011) Auxin signaling and transport promote susceptibility to the root-infecting fungal pathogen Fusarium oxysporum in Arabidopsis. Mol Plant Microbe Interact 24:733–748
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36
Li B, Meng X, Shan L, He P (2016) Transcriptional regulation of pattern-triggered immunity in plants. Cell Host Microbe 19:641–650
Li E, Wang G, Yang Y, Xiao J, Mao Z, Xie B (2015) Microscopic analysis of the compatible and incompatible interactions between Fusarium oxysporum f. sp. conglutinans and cabbage. Eur J Plant Pathol 141:597–609
Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16:319–331
Lv H, Fang Z, Yang L, Zhang Y, Wang Q, Liu Y, Zhuang M, Yang Y, Xie B, Liu B, Liu J, Kang J, Wang X (2014) Mapping and analysis of a novel candidate Fusarium wilt resistance gene FOC1 in Brassica oleracea. BMC Genomics 15:1094
Lyons R, Stiller J, Powell J, Rusu A, Manners JM, Kazan K (2015) Fusarium oxysporum triggers tissue-specific transcriptional reprogramming in Arabidopsis thaliana. PLoS ONE 10:e0121902
Marone D, Russo MA, Laidò G, De Leonardis AM, Mastrangelo AM (2013) Plant nucleotide binding site-leucine-rich repeat (NBS-LRR) genes: active guardians in host defense responses. Int J Mol Sci 14:7302–7326
Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15:809–834
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Murray SL, Ingle RA, Petersen LN, Denby KJ (2007) Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol Plant Microbe Interact 20:1431–1438
Oumouloud A, El-Otmani M, Chikh-Rouhou H, Claver AG, Torres RG, Perl-Treves R, Álvarez JM (2013) Breeding melon for resistance to Fusarium wilt: recent developments. Euphytica 192:155–169
Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150:1648–1655
Ploetz RC (2006) Fusarium wilt of banana is caused by several pathogens referred to as Fusarium oxysporum f. sp. cubense. Phytopathology 96:653–656
Pu Z, Ino Y, Kimura Y, Tago A, Shimizu M, Natsume S, Sano Y, Fujimoto R, Kaneko K, Shea DJ, Fukai E, Fuji S-I, Hirano H, Okazaki K (2016) Changes in the proteome of xylem sap in Brassica oleracea in response to Fusarium oxysporum stress. Front Plant Sci 7:31
Pu Z, Shimizu M, Zhang Y, Nagaoka T, Hayashi T, Hori H, Matsumoto S, Fujimoto R, Okazaki K (2012) Genetic mapping of a fusarium wilt resistance gene in Brassica oleracea. Mol Breed 30:809–818
Ren D, Liu Y, Yang KY, Han L, Mao G, Glazebrook J, Zhang S (2008) A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 105:5638–5643
Saeki N, Kawanabe T, Ying H, Shimizu M, Kojima M, Abe H, Okazaki K, Kaji M, Taylor JM, Sakakibara H, Peacock WJ, Dennis ES, Fujimoto R (2016) Molecular and cellular characteristics of hybrid vigour in a commercial hybrid of Chinese cabbage. BMC Plant Biol 16:45
Shimizu M, Fujimoto R, Ying H, Pu Z, Ebe Y, Kawanabe T, Saeki N, Taylor JM, Kaji M, Dennis ES, Okazaki K (2014) Identification of candidate genes for fusarium yellows resistance in Chinese cabbage by differential expression analysis. Plant Mol Biol 85:247–257
Shimizu M, Pu Z, Kawanabe T, Kitashiba H, Matsumoto S, Ebe Y, Sano M, Funaki T, Fukai E, Fujimoto R, Okazaki K (2015) Map-based cloning of a candidate gene conferring Fusarium yellows resistance in Brassica oleracea. Theor Appl Genet 128:119–130
Spoel SH, Dong X (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 12:89–100
Strawn MA, Marr SK, Inoue K, Inada N, Zubieta C, Wildermuth MC (2007) Arabidopsis isochorismate synthase functional in pathogen-induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J Biol Chem 282:5919–5933
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578
Ulloa M, Hutmacher RB, Davis RM, Wright SD, Percy RG, Marsh B (2006) Breeding for Fusarium wilt race 4 resistance in cotton under field and greenhouse conditions. J Cotton Sci 10:114–127
Walker JC (1930) Inheritance of Fusarium resistance in cabbage. J Agric Res 40:721–745
Wang D, Amornsiripanitch N, Dong X (2006) A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog 2:e123
Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X (2007) Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol 17:1784–1790
Wang Z, Gerstein M, Synder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, Huang S, Li X, Hua W, Wang J, Wang X, Freeling M, Pires JC, Paterson AH, Chalhoub B, Wang B, Hayward A, Sharpe AG, Park BS, Weisshaar B, Liu B, Li B, Liu B, Tong C, Song C, Duran C, Peng C, Geng C, Koh C, Lin C, Edwards D, Mu D, Shen D, Soumpourou E, Li F, Fraser F, Conant G, Lassalle G, King GJ, Bonnema G, Tang H, Wang H, Belcram H, Zhou H, Hirakawa H, Abe H, Guo H, Wang H, Jin H, Parkin IAP, Batley J, Kim JS, Just J, Li J, Xu J, Deng J, Kim JA, Li J, Yu J, Meng J, Wang J, Min J, Poulain J, Wang J, Hatakeyama K, Wu K, Wang L, Fang L, Trick M, Links MG, Zhao M, Jin M, Ramchiary N, Drou N, Berkman PJ, Cai Q, Huang Q, Li R, Tabata S, Cheng S, Zhang S, Zhang S, Huang S, Sato S, Sun S, Kwon SJ, Choi SR, Lee TH, Fan W, Zhao X, Tan X, Xu X, Wang Y, Qiu Y, Yin Y, Li Y, Du Y, Liao Y, Lim Y, Narusaka Y, Wang Y, Wang Z, Li Z, Wang Z, Xiong Z, Zhang Z (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1039
Xing M, Lv H, Ma J, Xu D, Li H, Yang L, Kang J, Wang X, Fang Z (2016) Transcriptome profiling of resistance to Fusarium oxysporum f. sp. conglutinans in cabbage (Brassica oleracea) roots. PLoS ONE 11:e0148048
Zhu QH, Stephen S, Kazan K, Jin G, Fan L, Taylor J, Dennis ES, Helliwell CA, Wang MB (2013) Characterization of the defense transcriptome responsive to Fusarium oxysporum-infection in Arabidopsis using RNA-seq. Gene 512:259–266
Acknowledgements
We thank Dr. Zijing Pu for excellent technical assistance. We also thank Dr. Qian-Hao Zhu and Dr. Ming-Bo Wang for providing us the data of DEGs in A. thaliana. This work was supported in part by a Grant-in-Aid for Young Scientists (B) (2478002) (JSPS) to R. Fujimoto, Research Fellowships of JSPS for Young Scientists (12J05450) to M. Shimizu, and by the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (26022A) and by the Matching Planner Program from Japan Science and Technology Agency, JST (MP28116808421) to K. Okazaki.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Dr. Kinya Toriyama.
Electronic supplementary material
Below is the link to the electronic supplementary material.
299_2017_2198_MOESM1_ESM.pptx
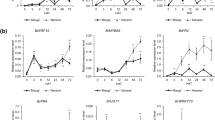
Figure S1. Gene expression levels of up-regulated genes after Foc inoculation measured by quantitative real-time RT-PCR. Six genes, BG3 (Group-2), Bra037877 (Group-1), GULLO2 (Group-2), JRG21 (Group-3), PDR12 (Group-2), and PRB1 (Group-2), whose expression was up-regulated in R-24 h-inf and/or S-24 h-inf, were examined. Expression levels of these six genes were measured by real-time RT-PCR at 6, 12, 24, and 72 h after Foc inoculation (HAI). Values are mean ± SE (three biological and technical replicates) of relative expression levels compared with Bractin were shown. NI, non-inoculated samples (PPTX 92 kb)
299_2017_2198_MOESM2_ESM.pptx
Figure S2. Validation of six genes at 24 HAI by quantitative real-time RT-PCR. The expression levels quantified by real-time RT-PCR and RNA-seq data at 24 HAI were compared in six genes, whose expression was up-regulated in Rb-inf and/or Sb-inf. Values are mean ± SE (three biological and technical replicates) of relative expression levels compared with Bractin are shown. The values of fragments per kilo-base per million (FPKM) were calculated by data from RNA-seq analysis. R-24 h-mock, mock-inoculated samples of the resistant line (RJKB-T23); S-24 h-mock; mock-inoculated samples of the susceptible line (RJKB-T24). R-24 h-inf, inoculated samples at 24 HAI of the resistant line; S-24 h-inf, inoculated samples at 24 HAI of the susceptible line; HAI, hours after inoculation. NI, non-inoculated samples (PPTX 105 kb)
299_2017_2198_MOESM3_ESM.pptx
Figure S3. Validation of six genes at 24 HAI by quantitative real-time RT-PCR. The expression levels quantified by real-time RT-PCR and RNA-seq data at 24 HAI were compared in six genes, whose expression was up-regulated in R-24 h-inf and/or S-24 h-inf. Values are mean ± SE (three biological and technical replicates) of relative expression levels compared with Bractin are shown. The values of fragments per kilo-base per million (FPKM) were calculated by data from RNA-seq analysis. R-24 h-mock, mock-inoculated samples of the resistant line (RJKB-T23); S-24 h-mock; mock-inoculated samples of the susceptible line (RJKB-T24). R-24 h-inf, inoculated samples at 24 HAI of the resistant line; S-24 h-inf, inoculated samples at 24 HAI of the susceptible line; HAI, hours after inoculation. NI, non-inoculated samples (PPTX 107 kb)
299_2017_2198_MOESM4_ESM.pptx
Figure S4. Validation of six genes at 72 HAI by quantitative real-time RT-PCR. The expression levels quantified by real-time RT-PCR and RNA-seq data at 72 HAI were compared in six genes, whose expression was up-regulated in Rb-inf and/or Sb-inf. Values are mean ± SE (three biological and technical replicates) of relative expression levels compared with Bractin are shown. The values of fragments per kilo-base per million (FPKM) were calculated by data from RNA-seq analysis. R-72 h-mock, mock-inoculated samples of the resistant line (RJKB-T23); S-72 h-mock; mock-inoculated samples of the susceptible line (RJKB-T24). R-72 h-inf, inoculated samples at 72 HAI of the resistant line; S-72 h-inf, inoculated samples at 72 HAI of the susceptible line; HAI, hours after inoculation. NI, non-inoculated samples (PPTX 100 kb)
299_2017_2198_MOESM5_ESM.pptx
Figure S5. Validation of six genes at 72 HAI by quantitative real-time RT-PCR. The expression levels quantified by real-time RT-PCR and RNA-seq data at 72 HAI were compared in six genes, whose expression was up-regulated in R-24 h-inf and/or S-24 h-inf. Values are mean ± SE (three biological and technical replicates) of relative expression levels compared with Bractin are shown. The values of fragments per kilo-base per million (FPKM) were calculated by data from RNA-seq analysis. R-72 h-mock, mock-inoculated samples of the resistant line (RJKB-T23); S-72 h-mock; mock-inoculated samples of the susceptible line (RJKB-T24). R-72 h-inf, inoculated samples at 72 HAI of the resistant line; S-72 h-inf, inoculated samples at 72 HAI of the susceptible line; HAI, hours after inoculation. NI, non-inoculated samples (PPTX 103 kb)
299_2017_2198_MOESM6_ESM.pptx
Figure S6. Venn diagram of up- or down-regulated genes in inoculated samples of B. rapa (24 HAI) compared with A. thaliana (1 DPI). R-24 h-inf UP/DOWN, up-/down-regulated genes in the inoculated whole plants compared with genes in the mock-inoculated whole plants in resistant line (RJKB-T23) at 24 HAI (hours after inoculation). S-24 h-inf UP/DOWN, up-/down-regulated genes in the inoculated whole plants compared with genes in the mock-inoculated whole plants in the susceptible line (RJKB-T24) at 24 HAI. At UP/DOWN, up-/down-regulated genes in inoculated samples at 1 DPI (day-post-inoculation) compared with genes in mock-inoculated samples in A. thaliana (PPTX 59 kb)
299_2017_2198_MOESM7_ESM.pptx
Figure S7. Validation of four genes at 24 HAI by quantitative real-time RT-PCR. The expression levels quantified by real-time RT-PCR and RNA-seq data at 24 HAI were compared in four genes, whose expression was up-regulated at 24 HAI in resistant line of B. rapa and 1 DPI (day-post-inoculation) in A. thaliana. Values are mean ± SE (three biological and technical replicates) of relative expression levels compared with Bractin are shown. The values of fragments per kilo-base per million (FPKM) were calculated by data from RNA-seq analysis. R-24 h-mock, mock-inoculated samples of the resistant line (RJKB-T23); S-24 h-mock; mock-inoculated samples of the susceptible line (RJKB-T24). R-24 h-inf, inoculated samples at 24 HAI of the resistant line; S-24 h-inf, inoculated samples at 24 HAI of the susceptible line; HAI, hours after inoculation. NI, non-inoculated samples (PPTX 84 kb)
299_2017_2198_MOESM8_ESM.pptx
Figure S8. Validation of four genes at 72 HAI by quantitative real-time RT-PCR. The expression levels quantified by real-time RT-PCR and RNA-seq data at 72 HAI were compared in four genes, whose expression was up-regulated at 24 HAI in resistant line of B. rapa (RJKB-T23) and 1 DPI (day-post-inoculation) in A. thaliana. Values are mean ± SE (three biological and technical replicates) of relative expression levels compared with Bractin are shown. The values of fragments per kilo-base per million (FPKM) were calculated from the RNA-seq data analysis. R-72 h-mock, mock-inoculated samples of the resistant line (RJKB-T23); S-72 h-mock; mock-inoculated samples of the susceptible line (RJKB-T24). R-72 h-inf, inoculated samples at 72 HAI of the resistant line (RJKB-T23); S-72 h-inf, inoculated samples at 72 HAI of the susceptible line (RJKB-T24); HAI, hours after inoculation. NI, non-inoculated samples (PPTX 84 kb)
Rights and permissions
About this article
Cite this article
Miyaji, N., Shimizu, M., Miyazaki, J. et al. Comparison of transcriptome profiles by Fusarium oxysporum inoculation between Fusarium yellows resistant and susceptible lines in Brassica rapa L.. Plant Cell Rep 36, 1841–1854 (2017). https://doi.org/10.1007/s00299-017-2198-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-017-2198-9