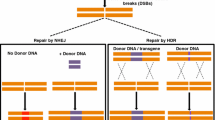
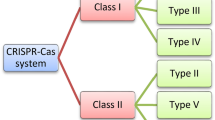
Abstract
The increasing burden of the world population on agriculture requires the development of more robust crops. Dissecting the basic biology that underlies plant development and stress responses will inform the design of better crops. One powerful tool for studying plants at the molecular level is the RNA-programmed genome editing system composed of a clustered regularly interspaced short palindromic repeats (CRISPR)-encoded guide RNA and the nuclease Cas9. Here, some of the recent advances in CRISPR/Cas9 technology that have profound implications for improving the study of plant biology are described. These tools are also paving the way towards new horizons for biotechnologies and crop development.




Similar content being viewed by others
References
Ali Z, Abulfaraj A, Idris A, Ali S, Tashkandi M, Mahfouz MM (2015) CRISPR/Cas9-mediated viral interference in plants. Genome Biol 16:238
Aouida M, Eid A, Ali Z, Cradick T, Lee C, Deshmukh H, Atef A, AbuSamra D, Gadhoum SZ, Merzaban J et al (2015) Efficient fdCas9 synthetic endonuclease with improved specificity for precise genome engineering. PLoS One 10:e0133373
Aragão FJL, Faria JC (2009) First transgenic geminivirus-resistant plant in the field. Nat Biotechnol 27:1086–1088 (author reply 1088–1089)
Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF (2014) DNA replicons for plant genome engineering. Plant Cell 26:151–163
Baltes NJ, Hummel AW, Konecna E, Cegan R (2015) Conferring resistance to geminiviruses with the CRISPR-Cas prokaryotic immune system. Nat Plants 1:15145
Bortesi L, Fischer R (2014) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 33:1–28
Carroll D (2011) Genome engineering with zinc-finger nucleases. Genetics 188:773–782
Cermak T, Baltes NJ, Čegan R, Zhang Y, Voytas DF (2015) High-frequency, precise modification of the tomato genome. Genome Biol 16:232
Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim J-S (2014) Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 24:132–141
Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF (2010) Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186:757–761
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Crosetto N, Mitra A, Silva MJ, Bienko M, Dojer N, Wang Q, Karaca E, Chiarle R, Skrzypczak M, Ginalski K et al (2013) Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods 10:361–365
Engler C, Gruetzner R, Kandzia R, Marillonnet S (2009) Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One 4:e5553
Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM (2013) Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods 10:1116–1121
Fauser F, Roth N, Pacher M, Ilg G (2012) In planta gene targeting. Proc Nat Acad Sci 109:7535–7540
Fauser F, Schiml S, Puchta H (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79:348–359
Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, Wang Z, Zhang Z, Zheng R, Yang L et al (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci 111:4632–4637
Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31:822–826
Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK (2014) Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32:279–284
Fujita T, Fujii H (2013) Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (eChIP) using CRISPR. Biochem Biophys Res Commun 439:1–25
Gao Y, Zhao Y (2014) Self-processing of ribozyme-flanked RNAs into guide RNAs in vitroand in vivofor CRISPR-mediated genome editing. J Integr Plant Biol 56:343–349
Gelvin SB (2003) Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev 67:16–37
Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345
Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC et al (2014) Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159:647–661
Guilinger JP, Thompson DB, Liu DR (2014) Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol 32:577–582
Gutierrez C (1999) Geminivirus DNA replication. Cell Mol Life Sci 56:313–329
Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA (2015) Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33:510–517
Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34:733–739
Honda A, Hirose M, Sankai T, Yasmin L, Yuzawa K, Honsho K, Izu H, Iguchi A, Ikawa M, Ogura A (2015) Single-step generation of rabbits carrying a targeted allele of the tyrosinase gene using CRISPR/Cas9. Exp Anim 64:31–37
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O et al (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31:827–832
Jayathilaka K, Sheridan SD, Bold TD, Bochenska K, Logan HL, Weichselbaum RR, Bishop DK, Connell PP (2008) A chemical compound that stimulates the human homologous recombination protein RAD51. Proc Natl Acad Sci USA 105:15848–15853
Ji X, Zhang H, Zhang Y, Wang Y, Gao C (2015) Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in plants. Nat Plants 1:1–5
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Keith Joung J (2016) High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529:490–495
Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H et al (2015) Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517:583–588
Krouk G, Lingeman J, Colon AM, Coruzzi G, Shasha D (2013) Gene regulatory networks in plants: learning causality from time and perturbation. Genome Biol 14:123
Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B (2011) TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res 39:359–372
Li J-F, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J (2013a) Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31:688–691
Li J, Blue R, Zeitler B, Strange TL, Pearl JR, Huizinga DH, Evans S, Gregory PD, Urnov FD, Petolino JF (2013b) Activation domains for controlling plant gene expression using designed transcription factors. Plant Biotechnol J 11:671–680
Li Z, Liu Z-B, Xing A, Moon BP, Koellhoffer JP, Huang L, Ward RT, Clifton E, Falco SC, Cigan AM (2015) Cas9-Guide RNA directed genome editing in soybean. Plant Physiol 169:960–970
Lowder LG, Zhang D, Baltes NJ, Paul JW, Tang X, Zheng X, Voytas DF, Hsieh T-F, Zhang Y, Qi Y (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol 169:971–985
Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T (2015a) Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci 112:3002–3007
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y et al (2015b) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8:1274–1284
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013a) RNA-guided human genome engineering via Cas9. Science 339:823–826
Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM (2013b) CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 31:833–838
Mansoor S, Zafar Y, Briddon RW (2006) Geminivirus disease complexes: the threat is spreading. Trends Plant Sci 11:209–212
Mao Y, Zhang Z, Feng Z, Wei P, Zhang H, Botella JR, Zhu JK (2015) Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol J 14:519–532
Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL (2015) Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol 33:538–542
Mikami M, Toki S, Endo M (2016) Precision targeted mutagenesis via Cas9 paired nickases in rice. Plant Cell Physiol. doi:10.1093/pcp/pcw049 (epub ahead of print)
Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ et al (2010) A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29:143–148
Moffat AS (1999) Geminiviruses emerge as serious crop threat. Science 286:1835
Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31:691–693
Nishizawa-Yokoi A, Cermak T, Hoshino T, Sugimoto K, Saika H, Mori A, Osakabe K, Hamada M, Katayose Y, Starker C et al (2015) A defect in DNA ligase 4 enhances the frequency of TALEN-mediated targeted mutagenesis in rice. Plant Physiol 170:653–666
Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK (2014) Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell 54:698–710
Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR (2013) High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 31:839–843
Pawlowski WP, Somers DA (1998) Transgenic DNA integrated into the oat genome is frequently interspersed by host DNA. Proc Natl Acad Sci 95:12106–12110
Petolino JF, Davies JP (2013) Designed transcriptional regulators for trait development. Plant Sci 201–202:128–136
Piatek A, Ali Z, Baazim H, Li L, Abulfaraj A, Al-Shareef S, Aouida M, Mahfouz MM (2015) RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol J 13:578–589
Puchta H (1999) Double-strand break-induced recombination between ectopic homologous sequences in somatic plant cells. Genetics 152:1173–1181
Puchta H (2016) Using CRISPR/Cas in three dimensions: towards synthetic plant genomes, transcriptomes and epigenomes. Plant J. doi:10.1111/tpj.13100 (epub ahead of print)
Puchta H, Dujon B, Hohn B (1996) Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc Natl Acad Sci 93:5055–5060
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA (2013a) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183
Qi Y, Zhang Y, Zhang F, Baller JA, Cleland SC, Ryu Y, Starker CG, Voytas DF (2013b) Increasing frequencies of site-specific mutagenesis and gene targeting in Arabidopsis by manipulating DNA repair pathways. Genome Res 23:547–554
Ran FA, Hsu PD, Lin C-Y, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y et al (2013) Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154:1380–1389
Sauer NJ, Narváez-Vásquez J, Mozoruk J, Miller RB, Warburg ZJ, Woodward MJ, Mihiret YA, Lincoln TA, Segami RE, Sanders SL et al (2016) Oligonucleotide-mediated genome editing provides precision and function to engineered nucleases and antibiotics in plants. Plant Physiol 170:1917–1928
Schiml S, Fauser F, Puchta H (2014) The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J 80:1139–1150
Shaked H, Melamed-Bessudo C, Levy AA (2005) High-frequency gene targeting in Arabidopsis plants expressing the yeast RAD54 gene. Proc Natl Acad Sci 102:12265–12269
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu J-L et al (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31:686–688
Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F (2016) Rationally engineered Cas9 nucleases with improved specificity. Science 351:84–88
Song J, Yang D, Xu J, Zhu T, Chen YE, Zhang J (2016) RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat Commun 7:10548
Steinert J, Schiml S, Fauser F, Puchta H (2015) Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J 84:1295–1305
Sun Y, Zhang X, Wu C, He Y, Ma Y, Hou H, Guo X, Du W, Zhao Y, Xia L (2016) Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol Plant 9:628–631
Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM (2015) Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol 169:931–945
Tiwari SB, Belachew A, Ma SF, Young M, Ade J, Shen Y, Marion CM, Holtan HE, Bailey A, Stone JK et al (2012) The EDLL motif: a potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J 70:855–865
Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK (2014) Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol 32:569–576
Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP et al (2015) GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33:187–197
Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD (2010) Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11:636–646
Wang C, Shen L, Fu Y, Yan C, Wang K (2015a) A simple CRISPR/Cas9 system for multiplex genome editing in rice. J Genet Genomics 42:703–706
Wang Z-P, Xing H-L, Dong L, Zhang H-Y, Han C-Y, Wang X-C, Chen Q-J (2015b) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16:1–12
Wiedenheft B, Sternberg SH, Doudna JA (2012) RNA-guided genetic silencing systems in bacteria and archaea. Nature 482:331–338
Woo JW, Kim J, Kwon SI, Corvalán C, Cho SW, Kim H, Kim S-G, Kim S-T, Choe S, Kim J-S (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33:1162–1164
Xie K, Minkenberg B, Yang Y (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA 112:3570–3575
Xing H-L, Dong L, Wang Z-P, Zhang H-Y, Han C-Y, Liu B, Wang X-C, Chen Q-J (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14:327
Yang C-F, Chen K-C, Cheng Y-H, Raja JAJ, Huang Y-L, Chien W-C, Yeh S-D (2014) Generation of marker-free transgenic plants concurrently resistant to a DNA geminivirus and a RNA tospovirus. Sci Rep 4:5717
Yin K, Han T, Liu G, Chen T, Wang Y, Yu AYL, Liu Y (2015) A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci Rep 5:14926
Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, Liu H, La Russa M, Xie M, Ding S et al (2015) Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 16:142–147
Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS et al (2015) Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160:339–350
Zetsche B, Volz SE, Zhang F (2015) A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol 33:139–142
Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N et al (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12:797–807
Zhang Y, Rajan R, Seifert HS, Mondragón A, Sontheimer EJ (2015a) DNase H activity of Neisseria meningitidis Cas9. Mol Cell 60:242–255
Zhang Z, Mao Y, Ha S, Liu W, Botella JR, Zhu JK (2015b) A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep. doi:10.1007/s00299-015-1900-z (epub ahead of print)
Acknowledgments
Due to limited space, we could not cite the entirety of the current literature that may also be important. We thank A. Malzahn for critical reading of the manuscript and thoughtful advice on its composition and content. This work is supported by startup funds from East Carolina University and a Collaborative Funding Grant (2016-CFG-8003) from North Carolina Biotechnology Center and Syngenta to YQ.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict interests.
Additional information
Communicated by T. Cardi.
Rights and permissions
About this article
Cite this article
Paul, J.W., Qi, Y. CRISPR/Cas9 for plant genome editing: accomplishments, problems and prospects. Plant Cell Rep 35, 1417–1427 (2016). https://doi.org/10.1007/s00299-016-1985-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-016-1985-z