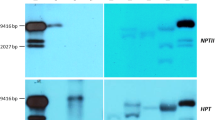
Abstract
To understand the genetic and molecular mechanisms underlying floral development in Populus tomentosa, we isolated PtLFY, a LEAFY homolog, from a P. tomentosa floral bud cDNA library. DNA gel blot analysis showed that PtLFY is present as a single copy in the genomes of both male and female individuals of P. tomentosa. The genomic copy is composed of three exons and two introns. Relative expression levels of PtLFY in tissues of P. tomentosa were estimated by RT-PCR; our results revealed that PtLFY mRNA is highly abundant in roots and both male and female floral buds. A low level of gene expression was detected in stems and vegetative buds, and no PtLFY-specific transcripts were detected in leaves. PtLFY expression patterns were analyzed during the development of both male and female floral buds in P. tomentosa via real-time quantitative RT-PCR. Continuous, stable and high-level expression of PtLFY-specific mRNA was detected in both male and female floral buds from September 13th to February 25th, but the level of PtLFY transcripts detected in male floral buds was considerably higher than in female floral buds. Our results also showed an inverted repeat PtLFY fragment (PtLFY-IR) effectively blocked flowering of transgenic tobacco plants, and that this effect appeared to be due to post-transcriptional silencing of the endogenous tobacco LFY homologs NFL1 and NFL2.






Similar content being viewed by others
Abbreviations
- E. coli :
-
Escherichia coli
- SDS:
-
Sodium dodecyl sulfate
- SSC:
-
Saline-sodium citrate
- WT:
-
Wild type
- qRT-PCR:
-
Quantitative reverse transcription polymerase chain reaction
- IR:
-
Inverted repeat
- PTGS:
-
Post-transcriptional gene silencing
References
Ahearn KP, Johnson HA, Weigel D, Wagner DR (2001) NFL1, a Nicotiana tabacum LEAFY-like gene, controls meristem initiation and floral structure. Plant Cell Physiol 42:1130–1139
Almada R, Cabrera N, Casaretto JA, Ruiz-Lara S, González Villanueva E (2009) VvCO and VvCOL1, two CONSTANS homologous genes, are regulated during flower induction and dormancy in grapevine buds. Plant Cell Rep 28(8):1193–1203
Blazquez MA, Weigel D (2000) Integration of floral inductive signals in Arabidopsis. Nature 404:889–892
Blazquez MA, Soowal LN, Lee I, Weigel D (1997) LEAFY expression and flower initiation in Arabidopsis. Development 124:3835–3844
Blázquez MA, Santos E, Flores CL, Martínez-Zapater JM, Salinas J, Gancedo C (1998) Isolation and molecular characterization of the Arabidopsis TPS1 gene, encoding trehalose-6-phosphate synthase. Plant J 13:685–689
Boes TK, Strauss SH (1994) Floral phenology and morphology of black cottonwood, Populus trichocarpa (Salicaceae). Am J Bot 81:562–567
Böhlenius H (2007) Control of flowering time and growth Cessation in Arabidopsis and Populus trees. Doctoral Thesis, Swedish University of Agricultural Sciences, Umeå, Sweden
Bomblies K, Wang R-L, Ambrose BA, Schmidt RJ, Meeley RB, Doebley J (2003) Duplicate FLORICAULA/LEAFY homologues zfl1 and zfl2 control inflorescence architecture and flower patterning in maize. Dev 130:2385–2395
Boss PK, Bastow RM, Mylne JS, Dean C (2004) Multiple pathways in the decision to flower enabling, promoting, and resetting. Plant Cell 16(Suppl):S18–S31
Braatne JH, Rood SB, Heilman PE (1996) Life history, ecology and conservation of riparian cottonwoods in North America. In: Stettler RF, Bradshaw HD, Heilman PE, Hinckley TM (eds) Populus and its implications for management and conservation, part I, chap 3. NRC Research Press, National Research Council of Canada, Ottawa, pp 57–85
Bradley D, Ratcliffe OJ, Vincent C, Carpenter R, Coen ES (1997) Inflorescence commitment and architecture in Arabidopsis. Science 275:80–83
Carmona MJ, Cubas P, Martinez-Zapater JM (2002) VFL, the Grapevine FLORICAULA/LEAFY ortholog, is expressed in meristematic regions independently of their fate. Plant Physiol 130:68–77
Chae E, Tan QK-G, Hill TA, Irish VF (2008) An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development 135:1235–1245
Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine tree. Plant Mol Biol Rep 11:113–116
Chuang CF, Meyerowitz EM (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA 97:4985–4990
Coen ES, Romero JM, Doyle S, Elliott R, Murphy G, Carpenter R (1990) Floricaula: a homeotic gene required for flower development in Antirrhinum majus. Cell 63:1311–1322
Dornelas MC, Rodriguez APM (2005) The rubber tree (Hevea brasiliensis Muell. Arg.) homologue of the LEAFY/FLORICAULA gene is preferentially expressed in both male and female floral meristems. J Exp Bot 56:1965–1974
Dornelas MC, Amaral WAN, Rodriguez APM (2004) EgLFY, the Eucalyptus grandis homolog of the Arabidopsis gene LFY is expressed in reproductive and vegetative tissues. Braz J Plant Physiol 16:105–114
Frohlich MW, Parker DS (2000) The mostly male theory of flower evolution origins: from genes to fossils. Sys Bot 25:155–170
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788
Hamès C, Ptchelkine D, Grimm C, Thevenon E, Moyroud E, Gérard F, Martiel JL, Benlloch R, Parcy F, Müller CW (2008) Structural basis for LEAFY floral switch function and similarity with helix-turn-helix proteins. EMBO J 27:2628–2637
Hsu CY, Liu YX, Luthe DS, Yuceer C (2006) Poplar FT2 shortens the juvenile phase and promotes seasonal flowering. Plant Cell 18:1846–1861
Junko K, Saeko K, Keisuke N, Takeshi I, Ko S (1998) Down-regulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation. PNAS 95:1979–1982
Kerschen A, Napoli CA, Jorgensen RA, Müller AE (2004) Effectiveness of RNA interference in transgenic plants. FEBS Lett 566:223–228
Kotoda N, Wada M, Kusaba S, Kano-Murakami Y, Masuda T, Soejima J (2002) Overexpression of dMADS5, an APETALA1-like gene of apple, causes early flowering in transgenic Arabidopsis. Plant Sci 162:679–687
Li Y, Hagen G, Guilfoyle TJ (1992) Altered morphology in transgenic tobacco plants that overproduce cytokinins in specific tissues and organs. Dev Biol 153:386–395
Lohmann JU, Weigel D (2002) Building beauty: the genetic control of floral patterning. Dev Cell 2:135–142
Maizel A, Busch MA, Tanahashi T, Perkovic J, Kato M, Hasebe M, Weigel D (2005) The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science 308:260–263
Mandel MA, Yanofsky MF (1995) A gene triggering flower formation in Arabidopsis. Nature 377:522–524
Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360:273–277
Mellerowicz EJ, Horgan K, Walden A, Coker A, Walter C (1998) PRFLL: a Pinus radiata homologue of FLORICAULA and LEAFY is expressed in buds containing vegetative and undifferentiated male cone primordia. Planta 206:619–629
Mouradov A, Glassick T, Hamdorf B, Murphy L, Fowler B, Marla S, Teasdale RD (1998) NEEDLY, a Pinus radiata ortholog of FLORICAULA/LEAFY genes, expressed in both reproductive and vegetative meristems. PNAS 95:6537–6542
Ng M, Yanofsky MF (2001) Function and evolution of the plant MADS-box gene family. Nat Rev Genet 2:186–195
Nilsson O, Lee I, Blazquez MA, Weigel D (1998) Flowering time genes modulate the response to LEAFY activity. Genetics 150:403–410
Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53(4):674–690
Peña L, Martin-Trillo MM, Juarez J, Pina JA, Navarro L, Martinez-Zapater JM (2001) Constitutive expression of Arabidopsis LEAFY or APETALA1 genes in citrus reduces their generation time. Nat Biotechnol 19:263–267
Reinhardt D, Kuhlemeiera C (2002) Plant architecture. EMBO Rep 3:846–851
Rottmann WH, Meilan R, Sheppard LA, Brunner AM, Skinner JS, Ma C, Cheng S, Jouanin L, Pilate G, Strauss SH (2000) Diverse effects of overexpression of LEAFY and PTLF, a poplar (Populus) homolog of LEAFY/FLORICAULA, in transgenic poplar and Arabidopsis. Plant J 22:235–245
Ruan YL, Llewellyn DJ, Furbank RT (2003) Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 15(4):952–964
Sheppard LA (1997) PTD: a Populus trichocarpa gene with homology to floral homeotic transcription factors. PhD Dissertation, Oregon State University, Corvallis, USA
Soltis DE, Soltis PS (1999) Polyploidy: recurrent formation and genome evolution. Trends Ecol Evol 14:348–352
Somerville C, Koornneef M (2002) A fortunate choice: the history of Arabidopsis as a model plant. Nat Rev Genet 3:883–889
Souer E, van der Krol A, Kloos D, Spelt C, Bliek M, Mol J, Koes R (1998) Genetic control of branching pattern and floral identity during Petunia inflorescence development. Development 125:733–742
Southerton SG, Strauss SH, Olive MR, Harcourt RL, Decroocq V, Zhu X, Llewellyn DJ, Peacock WJ, Dennis ES (1998) Eucalyptus has a functional equivalent of the Arabidopsis floral meristem identity gene LEAFY. Plant Mol Biol 37:897–910
Sunkar R, Jagadeeswaran G (2008) In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol 8:37
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tzfira T, Jensen CS, Wang WG, Zuker A, Vinocur B, Altman A, Vainstein A (1997) Transgenic Populus tremula: a step-by-step protocol for its Agrobacterium-mediated transformation. Plant Mol Biol Rep 15:219–235
Wada M, Cao Q-F, Kotoda N, Soejima J-I, Masuda T (2002) Apple has two orthologues of FLORICAULA/LEAFY involved in flowering. Plant Mol Biol 49:567–577
Walton EF, Podivinsky E, Wu RM (2001) Bimodal pattern of floral gene expression over the two seasons that kiwifruit flowers develop. Physiol Plantarum 111:396–404
Wei H, Meilan R, Brunner AM, Skinner JS, Ma C, Strauss SH (2006) Transgenic sterility in Populus: expression properties of the poplar PTLF, Agrobacterium NOS and two minimal 35S promoters in vegetative tissues. Tree Physiol 26(4):401–410
Weigel D, Meyerowitz EM (1993) Activation of floral homeotic genes in Arabidopsis. Science 261:1723–1726
Weigel D, Meyerowitz EM (1994) The ABCs of floral homeotic genes. Cell 78:203–209
Weigel D, Nilsson O (1995) A developmental switch sufficient for flower initiation in diverse plants. Nature 377:495–500
Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69:843–859
Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, Robinson SP, Gleave AP, Green AG, Waterhouse PM (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27:581–590
Wikström N, Savolainen V, Chase MW (2001) Evolution of the angiosperms: calibrating the family tree. Proc R Soc Lond B 268:2211–2220
Yanofsky MF (1995) Floral meristems to floral organs: Genes controlling early events in Arabidopsis flower development. Annu Rev Plant Physiol Plant Mol 46:167–188
Yuceer C, Land SB Jr, Kubiske ME, Harkess RL (2003) Shoot morphogenesis associated with flowering in Populus deltoides (Salicaceae). Am J Bot 90:196–206
Zhang Q, Zhang ZY, Lin SZ, Zheng HQ, Lin YZ, An XM, Li Y, Li HX (2008) Characterization of resistance gene analogs with a nucleotide binding site isolated from a triploid white poplar. Plant Biol (Stuttg) 10(3):310–322
Zheng H, Lin S, Zhang Q, Lei Y, Zhang Z (2009) Functional analysis of 5′ untranslated region of a TIR-NBS-enconding gene from triploid white poplar. Mol Genet Genomics 282(4):381–394
Acknowledgments
We gratefully acknowledge Prof. Phillip F. Elliott, East Hartford University, CT, USA, and Dr. Jean W. H. Yong, Nanyang Technological University, Singapore, for invaluable advice and assistance. We thank Huiquan Zheng, a Ph.D. student currently in our laboratory, for assistance with figure preparation. This work was supported by grants from the Natural Science Foundation of China (No. 30571511), National High-tech R&D Program of China (No. 2009AA10Z107) and Key Project of Education of Chinese Ministry of Education (No. 108017).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by B. Li.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
The predicted secondary and tertiary structures of the PtLFY protein. (A) The secondary structure of PtLFY. The blue, yellow, green and light blue colors indicate the helix, sheet, turn and coil structure, respectively. (B) Putative proportion of helix, sheet, turn and coil in PtLFY. (C) The tertiary structure of PtLFY. The highly conserved PtLFY C-terminal is composed of α1, α2, α3, α4, α5, α6 and α7 helices (TIFF 4662 kb)
Fig. S2
The alignments of C-terminal amino acid sequences (A) and nucleotide sequences (B) of PtLFY (GenBank accession no. AY211519) with NFL1 (U16172) and NFL2 (U15799). PtLFY shares 94.7% amino acid identity with NFL1 and NFL2 at C-terminal designed as the target of PtLFY-IR structure (A). Nucleic acid of PtLFY shows 82.1% similarity to that of NFL1 and NFL2 at C-terminal designed as the target of PtLFY-IR structure (B) (TIFF 1128 kb)
Rights and permissions
About this article
Cite this article
An, XM., Wang, DM., Wang, ZL. et al. Isolation of a LEAFY homolog from Populus tomentosa: expression of PtLFY in P. tomentosa floral buds and PtLFY-IR-mediated gene silencing in tobacco (Nicotiana tabacum). Plant Cell Rep 30, 89–100 (2011). https://doi.org/10.1007/s00299-010-0947-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-010-0947-0