Abstract
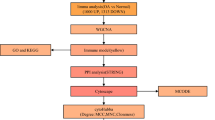
Osteoarthritis (OA) is a primary leading cause of pain and disability. However, some cases are diagnosed at the later stage which delayed the timely treatment. This study aims to identify effective diagnostic signature for OA. The mRNA profile GSE48566 including 106 blood samples of OA patients and 33 blood samples of healthy individuals was downloaded from Gene Expression Omnibus (GEO) database. The potential OA-related genes were screened by weighted gene co-expression network analysis (WGCNA). Gene ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed to reveal the functions or pathways of OA-related genes using the clusterProfiler function package of R software. Key genes significantly involved in OA progression were further screened by protein–protein interaction (PPI) network. The logistic regression model and the random forest model were conducted by bringing into optimal genes selected by stepwise regression analysis, and fivefold cross validation method was used to determine their reliability. A total of 146 genes, existed in three modules and might be associated with the occurrence of OA, were screened. 15 genes were screened from the PPI network and four genes, including CCR6, CLEC7A, IL18 and SRSF2, were further optimized. Finally, a logistic regression model and a random forest model were conducted by bringing into four optimal genes, and could reliably separate OA patients from healthy subjects. Our study established two effective diagnostic models based on CCR6, CLEC7A, IL18 and SRSF2, which could reliably separate OA patients from healthy subjects.




Similar content being viewed by others
Availability of data and materials
The mRNA profile GSE48566 consist of 106 blood samples from OA patients and 33 blood samples from healthy subjects was downloaded from Gene Expression Omnibus (GEO) database.
References
Martel-Pelletier J, Barr AJ, Cicuttini FM, Conaghan PG, Cooper C, Goldring MB, Goldring SR, Jones G, Teichtahl AJ, Pelletier JP (2016) Osteoarthritis. Nat Rev Dis Prim 2:16072. https://doi.org/10.1038/nrdp.2016.72
Neogi T, Zhang Y (2013) Epidemiology of osteoarthritis. Rheum Dis Clin N Am 39(1):1–19. https://doi.org/10.1016/j.rdc.2012.10.004
Reichmann WM, Maillefert JF, Hunter DJ, Katz JN, Conaghan PG, Losina E (2011) Responsiveness to change and reliability of measurement of radiographic joint space width in osteoarthritis of the knee: a systematic review. Osteoarthr Cartil 19(5):550–556. https://doi.org/10.1016/j.joca.2011.01.023
Camacho-Encina M, Balboa-Barreiro V, Rego-Perez I, Picchi F, VanDuin J, Qiu J, Fuentes M, Oreiro N, LaBaer J, Ruiz-Romero C, Blanco FJ (2019) Discovery of an autoantibody signature for the early diagnosis of knee osteoarthritis: data from the osteoarthritis initiative. Ann Rheum Dis 78(12):1699–1705. https://doi.org/10.1136/annrheumdis-2019-215325
Nelson AE (2018) Osteoarthritis year in review 2017: clinical. Osteoarthr Cartil 26(3):319–325. https://doi.org/10.1016/j.joca.2017.11.014
Tulay P, Temel SG, Ergoren MC (2019) Investigation of KCNQ1 polymorphisms as biomarkers for cardiovascular diseases in the Turkish Cypriots for establishing preventative medical measures. Int J Biol Macromol 124:537–540. https://doi.org/10.1016/j.ijbiomac.2018.11.227
Zhang YW, Ding LS, Lai MD (2003) Reg gene family and human diseases. World J Gastroenterol 9(12):2635–2641. https://doi.org/10.3748/wjg.v9.i12.2635
Shukla KK, Misra S, Pareek P, Mishra V, Singhal B, Sharma P (2017) Recent scenario of microRNA as diagnostic and prognostic biomarkers of prostate cancer. Urol Oncol 35(3):92–101. https://doi.org/10.1016/j.urolonc.2016.10.019
Ntoumou E, Tzetis M, Braoudaki M, Lambrou G, Poulou M, Malizos K, Stefanou N, Anastasopoulou L, Tsezou A (2017) Serum microRNA array analysis identifies miR-140-3p, miR-33b-3p and miR-671-3p as potential osteoarthritis biomarkers involved in metabolic processes. Clin Epigenet 9:127. https://doi.org/10.1186/s13148-017-0428-1
Li J, Lan CN, Kong Y, Feng SS, Huang T (2018) Identification and analysis of blood gene expression signature for osteoarthritis with advanced feature selection methods. Front Genet 9:246. https://doi.org/10.3389/fgene.2018.00246
Ramos YF, Bos SD, Lakenberg N, Bohringer S, den Hollander WJ, Kloppenburg M, Slagboom PE, Meulenbelt I (2014) Genes expressed in blood link osteoarthritis with apoptotic pathways. Ann Rheum Dis 73(10):1844–1853. https://doi.org/10.1136/annrheumdis-2013-203405
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinform 9:559. https://doi.org/10.1186/1471-2105-9-559
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV (2019) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47(D1):D607–D613. https://doi.org/10.1093/nar/gky1131
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Friedman J, Hastie T, Tibshirani R (2010) Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33(1):1–22
Hu YH, Tai CT, Tsai CF, Huang MW (2018) Improvement of adequate digoxin dosage: an application of machine learning approach. J Healthc Eng. https://doi.org/10.1155/2018/3948245
Du J, Wang S, He C, Zhou B, Ruan YL, Shou H (2017) Identification of regulatory networks and hub genes controlling soybean seed set and size using RNA sequencing analysis. J Exp Bot 68(8):1955–1972. https://doi.org/10.1093/jxb/erw460
Greenham K, Guadagno CR, Gehan MA, Mockler TC, Weinig C, Ewers BE, McClung CR (2017) Temporal network analysis identifies early physiological and transcriptomic indicators of mild drought in Brassica rapa. Elife. https://doi.org/10.7554/eLife.29655
Rychel JK (2010) Diagnosis and treatment of osteoarthritis. Top Companion Anim Med 25(1):20–25. https://doi.org/10.1053/j.tcam.2009.10.005
Berenbaum F (2013) Osteoarthritis as an inflammatory disease (osteoarthritis is not osteoarthrosis!). Osteoarthr Cartil 21(1):16–21. https://doi.org/10.1016/j.joca.2012.11.012
Woodell-May JE, Sommerfeld SD (2020) Role of inflammation and the immune system in the progression of osteoarthritis. J Orthop Res 38(2):253–257. https://doi.org/10.1002/jor.24457
Millerand M, Berenbaum F, Jacques C (2019) Danger signals and inflammaging in osteoarthritis. Clin Exp Rheumatol 37(Suppl 120):48–56
Alaaeddine N, Antoniou J, Moussa M, Hilal G, Kreichaty G, Ghanem I, Abouchedid W, Saghbini E, Di Battista JA (2015) The chemokine CCL20 induces proinflammatory and matrix degradative responses in cartilage. Inflamm Res 64(9):721–731. https://doi.org/10.1007/s00011-015-0854-5
Lisignoli G, Manferdini C, Codeluppi K, Piacentini A, Grassi F, Cattini L, Filardo G, Facchini A (2009) CCL20/CCR6 chemokine/receptor expression in bone tissue from osteoarthritis and rheumatoid arthritis patients: different response of osteoblasts in the two groups. J Cell Physiol 221(1):154–160. https://doi.org/10.1002/jcp.21839
Kapoor M, Martel-Pelletier J, Lajeunesse D, Pelletier JP, Fahmi H (2011) Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat Rev Rheumatol 7(1):33–42. https://doi.org/10.1038/nrrheum.2010.196
Vicenti G, Bizzoca D, Carrozzo M, Solarino G, Moretti B (2018) Multi-omics analysis of synovial fluid: a promising approach in the study of osteoarthritis. J Biol Regul Homeost Agents 32(6 Suppl. 1):9–13
Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, Rotter JI, Wang HL, McGovern DP, Brown GD, Underhill DM (2012) Interactions between commensal fungi and the C-type lectin receptor dectin-1 influence colitis. Science 336(6086):1314–1317. https://doi.org/10.1126/science.1221789
Wang Z, Li K, Chen W, Wang X, Huang Y, Wang W, Wu W, Cai Z, Huang W (2020) Modulation of SRSF2 expression reverses the exhaustion of TILs via the epigenetic regulation of immune checkpoint molecules. Cell Mol Life Sci 77(17):3441–3452. https://doi.org/10.1007/s00018-019-03362-4
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
Conceptualization, Methodology and investigation: WZ and WX; Data curation and Formal Analysis: QQ and BS; Writing—original draft preparation: WZ, WX, QQ and BS; Writing—review and editing: WX; All co-authors take full responsibility for all aspects of the work.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Consent for publication
All the co-authors agree to the publication of the article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
296_2021_4795_MOESM2_ESM.tif
Supplementary file2 The diagnosis diagram of logistic regression model. The diagram of Residuals vs. Leverage. The red dotted line indicates the COOK distance. Generally, a point with the COOK greater than 0.5 is a very “influentia” point, which affects the reliability of the model. (TIF 1897 KB)
296_2021_4795_MOESM3_ESM.tif
Supplementary file3 The distribution of mRNA expression values. The horizontal axis represents the samples and the vertical axis represent mRNA expression vaules. (TIF 9134 KB)
Rights and permissions
About this article
Cite this article
Zhang, W., Qiu, Q., Sun, B. et al. A four-genes based diagnostic signature for osteoarthritis. Rheumatol Int 41, 1815–1823 (2021). https://doi.org/10.1007/s00296-021-04795-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00296-021-04795-6