Abstract
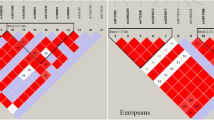
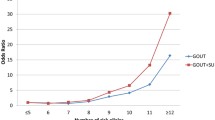
A genome-wide association study of gout in European populations identified 12 genetic variants strongly associated with risk of gout, but it is unknown whether these variants are also associated with gout risk in Chinese populations. A total of 145 patients with gout and 310 healthy control patients were recruited for a case–control association study. Twelve SNPs of CLNK and ZNF518B gene were genotyped, and association analysis was performed. Odds ratios (ORs) with 95 % confidence intervals (CIs) were used to assess the association. Overall, we found four risk alleles for gout in patients: the allele “G” of rs2041215 and rs1686947 in the CLNK gene by dominant model (OR 1.66; 95 % CI 1.04–2.63; p = 0.031) (OR 2.19; 95 % CI 1.38–3.46; p = 0.001) and additive model (OR 1.39; 95 % CI 1.00–1.93; p = 0.049) (OR 1.67; 95 % CI 1.19–2.32; p = 0.003), respectively, and the allele “A” of rs10938799 and rs10016022 in ZNF518B gene by recessive model (OR 4.66; 95 % CI 1.44–15.09; p = 0.008) (OR 4.54; 95 % CI 1.23–16.76; p = 0.020). Further haplotype analysis showed that the TCATTCTGA haplotype of CLNK was more frequent among patients with gout (adjusted OR 0.48; 95 % CI 0.24–0.95; p = 0.036). Additionally, polymorphisms of rs2041215, rs10938799, and rs17467273 were also correlated with clinical pathological parameters. This study provides evidence for gout susceptibility genes, CLNK and ZNF518B, in a Chinese population, which may have potential as diagnostic and prognostic marker for gout patients.


Similar content being viewed by others
References
Robinson PC, Horsburgh S (2014) Gout: joints and beyond, epidemiology, clinical features, treatment and co-morbidities. Maturitas 78(4):245–251
Roddy E, Doherty M (2010) Epidemiology of gout. Arthritis Res Ther 12(6):223
Wang SM, Aranda GA Jr, Gao S, Patel BV (2013) Benefit restrictions and gout treatment. J Manag Care Pharm JMCP 19(9):773–782
Sautner J (2014) Diagnosis and management of gout in Austria : survey of current practice considering the EULAR recommendations. Zeitschrift fur Rheumatologie. PubMed PMID: 24763908. Diagnose und Therapiestandard der Gicht in Osterreich : Umfrage unter Berucksichtigung der EULAR-Empfehlungen
Bardin T, Richette P (2011) The epidemiology and genetic of gout. Presse medicale. Epidemiologie et genetique de la goutte 40(9 Pt 1):830–835
Miao Z, Li C, Chen Y, Zhao S, Wang Y, Wang Z et al (2008) Dietary and lifestyle changes associated with high prevalence of hyperuricemia and gout in the Shandong coastal cities of Eastern China. J Rheumatol 35(9):1859–1864
Sakiyama M, Matsuo H, Shimizu S, Nakashima H, Nakayama A, Chiba T et al (2014) A common variant of organic anion transporter 4 (OAT4/SLC22A11) gene is associated with renal underexcretion type gout. Drug Metab Pharmacokinet 29(2):208–210
Vitale A, Cantarini L, Rigante D, Bardelli M, Galeazzi M (2014) Anakinra treatment in patients with gout and type 2 diabetes. Clin Rheumatol [Epub ahead of print]
Stamp LK, Chapman PT (2013) Gout and its comorbidities: implications for therapy. Rheumatology 52(1):34–44
Zhou L, Liu L, Liu X, Chen P, Zhang Y, Wu Y et al (2014) Systematic review and meta-analysis of the clinical efficacy and adverse effects of chinese herbal decoction for the treatment of gout. PLoS One 9(1):e85008
Roddy E, Choi HK (2014) Epidemiology of gout. Rheum Dis Clin North Am 40(2):155–175
Stark K, Reinhard W, Neureuther K, Wiedmann S, Sedlacek K, Baessler A et al (2008) Association of common polymorphisms in GLUT9 gene with gout but not with coronary artery disease in a large case–control study. PLoS One 3(4):e1948
Graessler J, Graessler A, Unger S, Kopprasch S, Tausche AK, Kuhlisch E et al (2006) Association of the human urate transporter 1 with reduced renal uric acid excretion and hyperuricemia in a German Caucasian population. Arthritis Rheum 54(1):292–300
Kolz M, Johnson T, Sanna S, Teumer A, Vitart V, Perola M et al (2009) Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet 5(6):e1000504
Wang J, Liu S, Wang B, Miao Z, Han L, Chu N et al (2012) Association between gout and polymorphisms in GCKR in male Han Chinese. Hum Genet 131(7):1261–1265
Kottgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C et al (2013) Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet. 45(2):145–154
Kochl S, Niederstatter H, Parson W (2005) DNA extraction and quantitation of forensic samples using the phenol–chloroform method and real-time PCR. Methods Mol Biol 297:13–30
Gabriel S, Ziaugra L, Tabbaa D (2009) SNP genotyping using the Sequenom MassARRAY iPLEX platform. In: Current protocols in human genetics, Chapter 2, Unit 2.12
Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM et al (2007) High-throughput oncogene mutation profiling in human cancer. Nat Genet 39(3):347–351
Adamec C (1964) [Example of the use of the nonparametric test. test X2 for comparison of 2 independent examples. Ceskoslovenske zdravotnictvi. 12:613–619. PubMed PMID: 14246305. Epub 1964/12/01. P r’iklad pou zit’i neparametrick’eho testu. text x2 pro srovn’an’i dvou nez’avisl’ych v’yb eru. cze
Bland JM, Altman DG (2000) Statistics notes. The odds ratio. BMJ 320(7247):1468
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81(3):559–575
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2):263–265
Shi YY, He L (2005) SHEsis a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 15(2):97–98
Goitsuka R, Kanazashi H, Sasanuma H, Fujimura YI, Hidaka Y, Tatsuno A et al (2000) A BASH/SLP-76-related adaptor protein MIST/Clnk involved in IgE receptor-mediated mast cell degranulation. Int Immunol 12(4):573–580
Cao MY, Davidson D, Yu J, Latour S, Veillette A (1999) Clnk, a novel SLP-76—related adaptor molecule expressed in cytokine-stimulated hemopoietic cells. J Exp Med 190(10):1527–1534
Darlington L, Scott J (1972) Plasma lipid levels in gout. Ann Rheum Dis 31(6):487
Wei WH, Guo Y, Kindt AS, Merriman TR, Semple CA, Wang K et al (2014) Abundant local interactions in the 4p16. 1 region suggest functional mechanisms underlying SLC2A9 associations with human serum uric acid. Hum Mol Genet 23(19):5061–5068
Acknowledgments
This work is supported by the Social Science Foundation of Chinese Ministry of Education (No. 12YJA850011), the Key Program of Natural Science Foundation of Xizang (Tibet) Autonomous Region (201122, 2014), and Major Training Program of Tibet University for Nationalities (No. 13myZP06). We would also like to thank BioScience Writers (BSW) for the assistance in the preparation of this manuscript.
Conflict of interest
The authors have no conflicts of interest to report.
Ethical standard
The use of samples was approved by the Human Research Committee of the Affiliated Hospital of Tibet University for Nationalities for Approval of Research Involving Human Subjects.
Author information
Authors and Affiliations
Corresponding author
Additional information
Tian-bo Jin and Yongchao Ren are joint first author.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jin, Tb., Ren, Y., Shi, X. et al. Genetic variations in the CLNK gene and ZNF518B gene are associated with gout in case–control sample sets. Rheumatol Int 35, 1141–1147 (2015). https://doi.org/10.1007/s00296-015-3215-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00296-015-3215-3