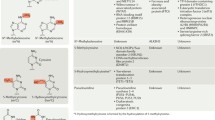
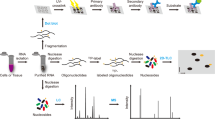
Abstract
Recent studies underscore RNA modifications as a novel mechanism to coordinate expression and function of different genes. While modifications on the sugar or base moieties of tRNA are well known, their roles in mRNA regulation are only starting to emerge. Interestingly, some modifications are present in both tRNA and mRNA, and here we discuss the functional significance of these common features. We describe key modifications that are present in both RNA types, elaborate on proteins that interact with them, and indicate recent works that identify roles in communicating tRNA processes and mRNA regulation. We propose that as tools are developed, the shortlist of features that are common between types of RNA will greatly expand and proteins that interact with them will be identified. In conclusion, the presence of the same modification in both RNA types provides an intersect between tRNA processes and mRNA regulation and implies a novel mechanism for connecting diverse cellular processes.


Similar content being viewed by others
Availability of data and material
All material is included in the manuscript.
Code availability
None.
References
Allnér O, Nilsson L (2011) Nucleotide modifications and tRNA anticodon-mRNA codon interactions on the ribosome. RNA 17:2177–2188. https://doi.org/10.1261/rna.029231.111
Ansman I, Motorin Y, Massenet S et al (2001) Identification and characterization of the tRNA: Ψ 31-synthase (Pus6p) of Saccharomyces cerevisiae. J Biol Chem 276:34934–34940. https://doi.org/10.1074/jbc.M103131200
Boccaletto P, MacHnicka MA, Purta E et al (2018) MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46:D303–D307. https://doi.org/10.1093/nar/gkx1030
Bohnsack KE, Höbartner C, Bohnsack MT (2019) Eukaryotic 5-methylcytosine (M 5 C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes (Basel) 10
Borchardt EK, Martinez NM, Gilbert WV (2020) Regulation and function of RNA pseudouridylation in human cells. Annu Rev Genet 54:309–336
Carlile TM, Rojas-Duran MF, Zinshteyn B et al (2014) Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515:143–146. https://doi.org/10.1038/nature13802
Carlile TM, Martinez NM, Schaening C et al (2019) mRNA structure determines modification by pseudouridine synthase 1. Nat Chem Biol 15:966–974. https://doi.org/10.1038/s41589-019-0353-z
Chan CTY, Pang YLJ, Deng W et al (2012) Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun 3:1–9. https://doi.org/10.1038/ncomms1938
Cheng QY, Xiong J, Ma CJ et al (2020) Chemical tagging for sensitive determination of uridine modifications in RNA. Chem Sci 11:1878–1891. https://doi.org/10.1039/c9sc05094a
Dev RR, Ganji R, Singh SP et al (2017) Cytosine methylation by DNMT2 facilitates stability and survival of HIV-1 RNA in the host cell during infection. Biochem J 474:2009–2026. https://doi.org/10.1042/BCJ20170258
Durant PC, Davis DR (1997) The effect of pseudouridine and pH on the structure and dynamics of the anticodon stem-loop of tRNA(Lys,3). Nucleic Acids Symp Ser, pp 56–57
Frye M, Jaffrey SR, Pan T et al (2016) RNA modifications: what have we learned and where are we headed? Nat Rev Genet 17:365–372
Ganem NS, Lamm AT (2017) A-to-I RNA editing–thinking beyond the single nucleotide. RNA Biol 14:1690–1694
Garin S, Levi O, Cohen B et al (2020) Localization and RNA binding of mitochondrial aminoacyl tRNA synthetases. Genes (Basel) 11:1–20
Gerber AP, Keller W (1999) An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 80(286):1146–1149. https://doi.org/10.1126/science.286.5442.1146
Gerber AP, Keller W (2001) RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem Sci 26:376–384
Goll MG, Kirpekar F, Maggert KA et al (2006) Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 80(311):395–398. https://doi.org/10.1126/science.1120976
Gregorova P, Sipari NH, Sarin LP (2020) Broad-range RNA modification analysis of complex biological samples using rapid C18-UPLC-MS. RNA Biol. https://doi.org/10.1080/15476286.2020.1853385
Han L, Phizicky EM (2018) A rationale for tRNA modification circuits in the anticodon loop. RNA 24:1277–1284
Heiss M, Hagelskamp F, Marchand V et al (2021) Cell culture NAIL-MS allows insight into human tRNA and rRNA modification dynamics in vivo. Nat Commun 12:389. https://doi.org/10.1038/s41467-020-20576-4
Helm M, Motorin Y (2017) Detecting RNA modifications in the epitranscriptome: predict and validate. Nat Rev Genet 18:275–291
Hopper AK, Nostramo RT (2019) TRNA processing and subcellular trafficking proteins multitask in pathways for other RNAs. Front Genet 10
Howell NW, Jora M, Jepson BF et al (2019) Distinct substrate specificities of the human tRNA methyltransferases TRMT10A and TRMT10B. RNA 25:1366–1376. https://doi.org/10.1261/rna.072090.119
Huang HY, Hopper AK (2016) Multiple layers of stress-induced regulation in tRNA biology. Life 6
Huang T, Chen W, Liu J et al (2019) Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat Struct Mol Biol 26:380–388. https://doi.org/10.1038/s41594-019-0218-x
Jordan Ontiveros R, Shen H, Stoute J et al (2020) Coordination of mRNA and tRNA methylations by TRMT10A. Proc Natl Acad Sci USA 117:7782–7791. https://doi.org/10.1073/pnas.1913448117
Khoddami V, Cairns BR (2013) Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol 31:458–464. https://doi.org/10.1038/nbt.2566
Kierzek E, Malgowska M, Lisowiec J et al (2014) The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res 42:3492–3501. https://doi.org/10.1093/nar/gkt1330
Levi O, Arava Y (2019) mRNA association by aminoacyl tRNA synthetase occurs at a putative anticodon mimic and autoregulates translation in response to tRNA levels. PLoS Biol 17:e3000274. https://doi.org/10.1371/journal.pbio.3000274
Levi O, Arava YS (2021) Pseudouridine-mediated translation control of mRNA by methionine aminoacyl tRNA synthetase. Nucleic Acids Res 49:432–443. https://doi.org/10.1093/nar/gkaa1178
Levi O, Garin S, Arava Y (2020) RNA mimicry in post-transcriptional regulation by aminoacyl tRNA synthetases. Wiley Interdiscip Rev RNA 11:e1564
Li X, Zhu P, Ma S et al (2015) Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol 11:592–597. https://doi.org/10.1038/nchembio.1836
Li X, Xiong X, Yi C (2016) Epitranscriptome sequencing technologies: decoding RNA modifications. Nat Methods 14:23–31
Liao S, Sun H, Xu C (2018) YTH domain: a family of N6-methyladenosine (m6A) readers. Genomics Proteomics Bioinform 16:99–107
Liddicoat BJ, Piskol R, Chalk AM et al (2015) RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 80(349):1115–1120. https://doi.org/10.1126/science.aac7049
Liu J, Yue Y, Han D et al (2014) A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10:93–95. https://doi.org/10.1038/nchembio.1432
Liu H, Begik O, Lucas MC et al (2019) Accurate detection of m6A RNA modifications in native RNA sequences. Nat Commun 10:1–9. https://doi.org/10.1038/s41467-019-11713-9
Lovejoy AF, Riordan DP, Brown PO (2014) Transcriptome-wide mapping of pseudouridines: Pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 9. https://doi.org/10.1371/journal.pone.0110799
Lyons SM, Fay MM, Ivanov P (2018) The role of RNA modifications in the regulation of tRNA cleavage. FEBS Lett 592:2828–2844. https://doi.org/10.1002/1873-3468.13205
Maity A, Das B (2016) N 6-methyladenosine modification in mRNA: machinery, function and implications for health and diseases. FEBS J 283:1607–1630. https://doi.org/10.1111/febs.13614
Marchand V, Pichot F, Neybecker P et al (2020) HydraPsiSeq: a method for systematic and quantitative mapping of pseudouridines in RNA. Nucleic Acids Res 48:e110. https://doi.org/10.1093/nar/gkaa769
Mathlin J, Le Pera L, Colombo T (2020) A census and categorization method of epitranscriptomic marks. Int J Mol Sci 21:4684. https://doi.org/10.3390/ijms21134684
Motorin Y, Lyko F, Helm M (2009) 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res 38:1415–1430. https://doi.org/10.1093/nar/gkp1117
Nishikura K (2010) Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem 79:321–349
O’Connell MA, Mannion NM, Keegan LP (2015) The epitranscriptome and innate immunity. PLOS Genet 11:e1005687. https://doi.org/10.1371/journal.pgen.1005687
Palladino MJ, Keegan LP, O’Connell MA, Reenan RA (2000) dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. RNA 6:1004–1018. https://doi.org/10.1017/S1355838200000248
Perez-Perri JI, Noerenberg M, Kamel W et al (2021) Global analysis of RNA-binding protein dynamics by comparative and enhanced RNA interactome capture. Nat Protoc 16:27–60. https://doi.org/10.1038/s41596-020-00404-1
Phizicky EM, Hopper AK (2010) tRNA biology charges to the front. Genes Dev 24:1832–1860
Picardi E, Manzari C, Mastropasqua F et al (2015) Profiling RNA editing in human tissues: towards the inosinome atlas. Sci Rep 5:14941. https://doi.org/10.1038/srep14941
Pinto Y, Levanon EY (2019) Computational approaches for detection and quantification of A-to-I RNA-editing. Methods 156:25–31
Rau K, Rösner L, Rentmeister A (2019) Sequence-specific m6A demethylation in RNA by FTO fused to RCas9. RNA 25:1311–1323. https://doi.org/10.1261/rna.070706.119
Rintala-Dempsey AC, Kothe U (2017) Eukaryotic stand-alone pseudouridine synthases—RNA modifying enzymes and emerging regulators of gene expression? RNA Biol 14:1185–1196. https://doi.org/10.1080/15476286.2016.1276150
Roundtree IA, He C (2016) RNA epigenetics—chemical messages for posttranscriptional gene regulation. Curr Opin Chem Biol 30:46–51
Roundtree IA, Evans ME, Pan T, He C (2017) Dynamic RNA modifications in gene expression regulation. Cell 169:1187–1200
Ru W, Zhang X, Yue B et al (2020) Insight into m6A methylation from occurrence to functions. Open Biol 10
Safra M, Nir R, Farouq D et al (2017) TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res 27:393–406. https://doi.org/10.1101/gr.207613.116
Schaefer M, Pollex T, Hanna K et al (2010) RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 24:1590–1595. https://doi.org/10.1101/gad.586710
Schwartz S, Bernstein DA, Mumbach MR et al (2014) Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159:148–162. https://doi.org/10.1016/j.cell.2014.08.028
Seeburg PH, Hartner J (2003) Regulation of ion channel/neurotransmitter receptor function by RNA editing. Curr Opin Neurobiol 13:279–283
Shanmugam R, Fierer J, Kaiser S et al (2015) Cytosine methylation of tRNA-Asp by DNMT2 has a role in translation of proteins containing poly-Asp sequences. Cell Discov 1:1. https://doi.org/10.1038/celldisc.2015.10
Shi H, Wei J, He C (2019) Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell 74:640–650
Tuorto F, Liebers R, Musch T et al (2012) RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol 19:900–905. https://doi.org/10.1038/nsmb.2357
Urdaneta EC, Vieira-Vieira CH, Hick T et al (2019) Purification of cross-linked RNA-protein complexes by phenol-toluol extraction. Nat Commun 10:1–17. https://doi.org/10.1038/s41467-019-08942-3
van Esveld SL, Spelbrink JN (2021) RNA crosslinking to analyze the mitochondrial RNA-binding proteome. In: Methods in molecular biology. Humana Press Inc, pp 147–158
Vilardo E, Amman F, Toth U et al (2020) Functional characterization of the human tRNA methyltransferases TRMT10A and TRMT10B. Nucleic Acids Res 48:6157–6169. https://doi.org/10.1093/nar/gkaa353
Wang X, Matuszek Z, Huang Y et al (2018) Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA 24:1305–1313. https://doi.org/10.1261/rna.067033.118
Xiong X, Yi C, Peng J (2017) Epitranscriptomics: toward a better understanding of RNA modifications. Genomics Proteomics Bioinform 15:147–153. https://doi.org/10.1016/j.gpb.2017.03.003
Xu L, Seki M (2020) Recent advances in the detection of base modifications using the nanopore sequencer. J Hum Genet 65:25–33
Yang X, Yang Y, Sun BF et al (2017) 5-methylcytosine promotes mRNA export-NSUN2 as the methyltransferase and ALYREF as an m 5 C reader. Cell Res 27:606–625. https://doi.org/10.1038/cr.2017.55
Zhang C, Fu J, Zhou Y (2019) A review in research progress concerning m6A methylation and immunoregulation. Front Immunol 10:922
Zheng G, Dahl JA, Niu Y et al (2013) ALKBH5 Is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49:18–29. https://doi.org/10.1016/j.molcel.2012.10.015
Zhou J, Wan J, Gao X et al (2015) Dynamic m6 a mRNA methylation directs translational control of heat shock response. Nature 526:591–594. https://doi.org/10.1038/nature15377
Acknowledgements
We thank Dr. Adi Golani-Armon for her critical reading of the manuscript and helpful suggestions. We apologize to many colleagues that we could not cite due to space limitations. Work in the lab is funded by ISF 258/18 and Michigan-Israel partnership. O.L. is recipient of the Jacobs fellowship for outstanding students.
Funding
Work in the lab is funded by ISF 258/18 and Michigan-Israel partnership. O.L. is recipient of the Jacobs fellowship for outstanding students.
Author information
Authors and Affiliations
Contributions
O.L. and Y.S. A. wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
None.
Additional information
Communicated by Michael Polymenis.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Levi, O., Arava, Y.S. RNA modifications as a common denominator between tRNA and mRNA. Curr Genet 67, 545–551 (2021). https://doi.org/10.1007/s00294-021-01168-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-021-01168-1