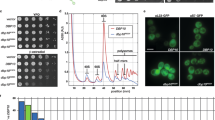
Abstract
Trypanosomatids are parasitic protozoans characterized by several unique structural and metabolic processes that include exquisite mechanisms associated with gene expression and regulation. During the initiation of protein synthesis, for instance, mRNA selection for translation seems to be mediated by different eIF4F-like complexes, which may play a significant role in parasite adaptation to different hosts. In eukaryotes, the heterotrimeric eIF4F complex (formed by eIF4E, eIF4G, and eIF4A) mediates mRNA recognition and ribosome binding and participates in various translation regulatory events. Six eIF4Es and five eIF4Gs have been described in trypanosomatids with several of these forming different eIF4F-like complexes. This has raised questions about their role in differential mRNA translation. Here we have studied further TbEIF4E2, the least known eIF4E homologue from Trypanosoma brucei, and found that it is not associated with an eIF4G homolog. It is, however, associated with mature mRNAs and binds to a histone mRNA stem-loop-binding protein (SLBP), one of two Trypanosoma SLBP homologs (TbSLBP1 and TbSLBP2). TbSLBP1 is more similar to the mammalian counterpart while TbSLBP2 is exclusive to trypanosomatids and related organisms. TbSLBP2 binds to TbEIF4E2 through a conserved central region missing in other SLBP homologs. Both SLBPs, as well as TbEIF4E2, were found to localize to the cytoplasm. TbEIF4E2 and TbSLBP2 are differentially expressed during cell culture, being more abundant in early-log phase, with TbSLBP2 also showing cell-cycle dependent expression. The new data reinforce unique aspects of the trypanosomatid eIF4Es, with the TbEIF4E2–TbSLBP complex possibly having a role in differential selection of mRNAs containing stem-loop structures.








Similar content being viewed by others
References
Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105. https://doi.org/10.1093/bioinformatics/bti263
Alsford S, Horn D (2004) Trypanosomatid histones. Mol Microbiol 53:365–372. https://doi.org/10.1111/j.1365-2958.2004.04151.x
Alsford S, Turner DJ, Obado SO et al (2011) High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res 21:915–924. https://doi.org/10.1101/gr.115089.110
Alsford S, duBois K, Horn D, Field MC (2012) Epigenetic mechanisms, nuclear architecture and the control of gene expression in trypanosomes. Expert Rev Mol Med 14:e13. https://doi.org/10.1017/erm.2012.7
Archer SK, Inchaustegui D, Queiroz R, Clayton C (2011) The cell cycle regulated transcriptome of Trypanosoma brucei. PLoS One 6:e18425. https://doi.org/10.1371/journal.pone.0018425
Aslett M, Aurrecoechea C, Berriman M et al (2010) TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res 38:D457–D462. https://doi.org/10.1093/nar/gkp851
Berriman M, Ghedin E, Hertz-Fowler C et al (2005) The genome of the African trypanosome Trypanosoma brucei. Science 309:416–422. https://doi.org/10.1126/science.1112642
Cakmakci NG, Lerner RS, Wagner EJ et al (2008) SLIP1, a factor required for activation of histone mRNA translation by the stem-loop binding protein. Mol Cell Biol 28:1182–1194. https://doi.org/10.1128/MCB.01500-07
Capewell P, Monk S, Ivens A et al (2013) Regulation of Trypanosoma brucei total and polysomal mRNA during development within its mammalian host. PLoS One 8:e67069. https://doi.org/10.1371/journal.pone.0067069
Chowdhury AR, Zhao Z, Englund PT (2008) Effect of hydroxyurea on procyclic Trypanosoma brucei: an unconventional mechanism for achieving synchronous growth. Eukaryot Cell 7:425–428. https://doi.org/10.1128/EC.00369-07
Clarkson BK, Gilbert WV, Doudna JA (2010) Functional overlap between eIF4G isoforms in Saccharomyces cerevisiae. PLoS One 5:e9114. https://doi.org/10.1371/journal.pone.0009114
Clayton CE (2002) Life without transcriptional control? From fly to man and back again. EMBO J 21:1881–1888. https://doi.org/10.1093/emboj/21.8.1881
Clayton C (2013) The regulation of trypanosome gene expression by RNA-binding proteins. PLoS Pathog 9:e1003680. https://doi.org/10.1371/journal.ppat.1003680
Clayton CE (2014) Networks of gene expression regulation in Trypanosoma brucei. Mol Biochem Parasitol 195:96–106. https://doi.org/10.1016/j.molbiopara.2014.06.005
Clayton C, Shapira M (2007) Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol Biochem Parasitol 156:93–101. https://doi.org/10.1016/j.molbiopara.2007.07.007
Culjkovic B, Topisirovic I, Skrabanek L et al (2006) eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol 175:415–426. https://doi.org/10.1083/jcb.200607020
Culjkovic B, Topisirovic I, Borden K (2007) Controlling gene expression through RNA regulons. Cell Cycle 6:65–69. https://doi.org/10.4161/cc.6.1.3688
Cunningham I (1977) New culture medium for maintenance of tsetse tissues and growth of trypanosomatids. J Protozool 24:325–329
Dávila López M, Samuelsson T (2008) Early evolution of histone mRNA 3′ end processing. RNA 14:1–10. https://doi.org/10.1261/rna.782308
de Melo Neto OP, da Costa Lima TDC, Xavier CC et al (2015) The unique Leishmania EIF4E4 N-terminus is a target for multiple phosphorylation events and participates in critical interactions required for translation initiation. RNA Biol 12:1209–1221. https://doi.org/10.1080/15476286.2015.1086865
de Melo Neto OPOP., Reis CRS, Moura DMNDMN et al (2016) Unique and conserved features of the protein synthesis apparatus in parasitic trypanosomatid (Trypanosoma and Leishmania) species. In: Hernández G, Jagus R (eds) Evolution of the protein synthesis machinery and its regulation. Springer International Publishing, Cham, pp 435–475
Dhalia R, Reis CRSRS., Freire ERER. et al (2005) Translation initiation in Leishmania major: characterisation of multiple eIF4F subunit homologues. Mol Biochem Parasitol 140:23–41. https://doi.org/10.1016/j.molbiopara.2004.12.001
Ebenezer TE, Carrington M, Lebert M et al (2017) Euglena gracilis genome and transcriptome: organelles, nuclear genome assembly strategies and initial features. Adv Exp Med Biol 979:125–140
Erben ED, Fadda A, Lueong S et al (2014) A genome-wide tethering screen reveals novel potential post-transcriptional regulators in Trypanosoma brucei. PLoS Pathog 10:e1004178. https://doi.org/10.1371/journal.ppat.1004178
Erkmann JA, Wagner EJ, Dong J et al (2005) Nuclear import of the stem-loop binding protein and localization during the cell cycle. Mol Biol Cell 16:2960–2971. https://doi.org/10.1091/mbc.E04-11-1023
Ersfeld K, Docherty R, Alsford S, Gull K (1996) A fluorescence in situ hybridisation study of the regulation of histone mRNA levels during the cell cycle of Trypanosoma brucei. Mol Biochem Parasitol 81:201–209
Estévez AM (2008) The RNA-binding protein TbDRBD3 regulates the stability of a specific subset of mRNAs in trypanosomes. Nucleic Acids Res 36:4573–4586. https://doi.org/10.1093/nar/gkn406
Fenn K, Matthews KR (2007) The cell biology of Trypanosoma brucei differentiation. Curr Opin Microbiol 10:539–546. https://doi.org/10.1016/j.mib.2007.09.014
Fernández-Moya SM, Estévez AM (2010) Posttranscriptional control and the role of RNA-binding proteins in gene regulation in trypanosomatid protozoan parasites. Wiley Interdiscip Rev RNA 1:34–46. https://doi.org/10.1002/wrna.6
Figueiredo LM, Cross GAM, Janzen CJ (2009) Epigenetic regulation in African trypanosomes: a new kid on the block. Nat Rev Microbiol 7:504–513. https://doi.org/10.1038/nrmicro2149
Florens L, Carozza MJ, Swanson SK et al (2006) Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 40:303–311. https://doi.org/10.1016/j.ymeth.2006.07.028
Freire ER, Dhalia R, Moura DMN et al (2011) The four trypanosomatid eIF4E homologues fall into two separate groups, with distinct features in primary sequence and biological properties. Mol Biochem Parasitol 176:25–36. https://doi.org/10.1016/j.molbiopara.2010.11.011
Freire ER, Malvezzi AM, Vashisht AA et al (2014a) Trypanosoma brucei translation initiation factor homolog EIF4E6 forms a tripartite cytosolic complex with EIF4G5 and a capping enzyme homolog. Eukaryot Cell 13:896–908. https://doi.org/10.1128/EC.00071-14
Freire ER, Vashisht AA, Malvezzi AM et al (2014b) eIF4F-like complexes formed by cap-binding homolog TbEIF4E5 with TbEIF4G1 or TbEIF4G2 are implicated in post-transcriptional regulation in Trypanosoma brucei. RNA 20:1272–1286. https://doi.org/10.1261/rna.045534.114
Freire E, Sturm N, Campbell D, de Melo Neto O (2017) The role of cytoplasmic mRNA cap-binding protein complexes in Trypanosoma brucei and other trypanosomatids. Pathogens 6:55. https://doi.org/10.3390/pathogens6040055
Furger A, Schürch N, Kurath U, Roditi I (1997) Elements in the 3′ untranslated region of procyclin mRNA regulate expression in insect forms of Trypanosoma brucei by modulating RNA stability and translation. Mol Cell Biol 17:4372–4380
De Gaudenzi J, Frasch AC, Clayton C (2005) RNA-binding domain proteins in kinetoplastids: a comparative analysis RNA-binding domain proteins in kinetoplastids: a comparative analysis. Society 4:2106–2114. https://doi.org/10.1128/EC.4.12.2106
Gazestani VH, Lu Z, Salavati R (2014) Deciphering RNA regulatory elements in trypanosomatids: one piece at a time or genome-wide? Trends Parasitol 30:234–240. https://doi.org/10.1016/j.pt.2014.02.008
Gingras AC, Raught B, Sonenberg N (1999) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68:913–963. https://doi.org/10.1146/annurev.biochem.68.1.913
Guindon S, Dufayard JF, Lefort V et al (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. https://doi.org/10.1093/sysbio/syq010
Günzl A, Kirkham JK, Nguyen TN et al (2015) Mono-allelic VSG expression by RNA polymerase I in Trypanosoma brucei: expression site control from both ends? Gene 556:68–73. https://doi.org/10.1016/j.gene.2014.09.047
Hill KL, Hutchings NR, Russell DG, Donelson JE (1999) A novel protein targeting domain directs proteins to the anterior cytoplasmic face of the flagellar pocket in African trypanosomes. J Cell Sci 112 Pt 18:3091–3101
Jackson RJ, Hellen CUT, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11:113–127. https://doi.org/10.1038/nrm2838
Jaeger S, Eriani G, Martin F (2004) Critical residues for RNA discrimination of the histone hairpin binding protein (HBP) investigated by the yeast three-hybrid system. FEBS Lett 556:265–270
Jaeger S, Barends S, Giegé R et al (2005) Expression of metazoan replication-dependent histone genes. Biochimie 87:827–834. https://doi.org/10.1016/j.biochi.2005.03.012
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Keene JD (2007) RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8:533–543. https://doi.org/10.1038/nrg2111
Kolev NG, Ramey-Butler K, Cross GAM et al (2012) Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science 338:1352–1353. https://doi.org/10.1126/science.1229641
Kolev NG, Ullu E, Tschudi C (2014) The emerging role of RNA-binding proteins in the life cycle of Trypanosoma brucei. Cell Microbiol 16:482–489. https://doi.org/10.1111/cmi.12268
Koseoglu MM, Graves LM, Marzluff WF (2008) Phosphorylation of threonine 61 by cyclin A/Cdk1 Triggers degradation of stem-loop binding protein at the end of S phase. Mol Cell Biol 28:4469–4479. https://doi.org/10.1128/MCB.01416-07
Kovtun O, Mureev S, Jung W et al (2011) Leishmania cell-free protein expression system. Methods 55:58–64. https://doi.org/10.1016/j.ymeth.2011.06.006
Kramer S (2012) Developmental regulation of gene expression in the absence of transcriptional control: the case of kinetoplastids. Mol Biochem Parasitol 181:61–72. https://doi.org/10.1016/j.molbiopara.2011.10.002
Kramer S, Queiroz R, Ellis L et al (2008) Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2(alpha) phosphorylation at Thr169. J Cell Sci 121:3002–3014. https://doi.org/10.1242/jcs.031823
Kramer S, Bannerman-Chukualim B, Ellis L et al (2013) Differential localization of the two T. brucei poly(A) binding proteins to the nucleus and RNP granules suggests binding to distinct mRNA pools. PLoS One 8:e54004. https://doi.org/10.1371/journal.pone.0054004
Labrecque R, Lodde V, Dieci C et al (2015) Chromatin remodelling and histone m RNA accumulation in bovine germinal vesicle oocytes. Mol Reprod Dev 82:450–462. https://doi.org/10.1002/mrd.22494
Lamb JR, Fu V, Wirtz E, Bangs JD (2001) Functional analysis of the trypanosomal AAA protein TbVCP with trans-dominant ATP hydrolysis mutants. J Biol Chem 276:21512–21520. https://doi.org/10.1074/jbc.M100235200
Lueong S, Merce C, Fischer B et al (2016) Gene expression regulatory networks in Trypanosoma brucei: insights into the role of the mRNA-binding proteome. Mol Microbiol 100:457–471. https://doi.org/10.1111/mmi.13328
MacGregor P, Matthews KR (2012) Identification of the regulatory elements controlling the transmission stage-specific gene expression of PAD1 in Trypanosoma brucei. Nucleic Acids Res 40:7705–7717. https://doi.org/10.1093/nar/gks533
Mader S, Lee H, Pause A, Sonenberg N (1995) The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol 15:4990–4997
Marzluff WF, Wagner EJ, Duronio RJ (2008) Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9:843–854. https://doi.org/10.1038/nrg2438
McKean PG (2003) Coordination of cell cycle and cytokinesis in Trypanosoma brucei. Curr Opin Microbiol 6:600–607. https://doi.org/10.1016/j.mib.2003.10.010
Moura DM, Reis CR, Xavier CC et al (2015) Two related trypanosomatid eIF4G homologues have functional differences compatible with distinct roles during translation initiation. RNA Biol 12:305–319. https://doi.org/10.1080/15476286.2015.1017233
Najafabadi HS, Lu Z, MacPherson C et al (2013) Global identification of conserved post-transcriptional regulatory programs in trypanosomatids. Nucleic Acids Res 41:8591–8600. https://doi.org/10.1093/nar/gkt647
Ouellette M, Papadopoulou B (2009) Coordinated gene expression by post-transcriptional regulons in African trypanosomes. J Biol 8:100. https://doi.org/10.1186/jbiol203
Peng J, Elias JE, Thoreen CC et al (2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC–MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res 2:43–50
Pereira MMC, Malvezzi AM, Nascimento LM et al (2013) The eIF4E subunits of two distinct trypanosomatid eIF4F complexes are subjected to differential post-translational modifications associated to distinct growth phases in culture. Mol Biochem Parasitol 190:82–86. https://doi.org/10.1016/j.molbiopara.2013.06.008
Polledo JM, Cervini G, Romaniuk MA, Cassola A (2016) Interactions between RNA-binding proteins and P32 homologues in trypanosomes and human cells. Curr Genet 62:203–212. https://doi.org/10.1007/s00294-015-0519-5
Ptushkina M, Malys N, McCarthy JEG (2004) eIF4E isoform 2 in Schizosaccharomyces pombe is a novel stress-response factor. EMBO Rep 5:311–316. https://doi.org/10.1038/sj.embor.7400088
Queiroz R, Benz C, Fellenberg K et al (2009) Transcriptome analysis of differentiating trypanosomes reveals the existence of multiple post-transcriptional regulons. BMC Genom 10:495. https://doi.org/10.1186/1471-2164-10-495
Raught B, Gingras AC (1999) eIF4E activity is regulated at multiple levels. Int J Biochem Cell Biol 31:43–57. https://doi.org/10.1016/S1357-2725(98)00131-9
Recinos RF, Kirchhoff LV, Donelson JE (2001) Cell cycle expression of histone genes in Trypanosoma cruzi. Mol Biochem Parasitol 113:215–222
Schimanski B, Nguyen TN, Günzl A (2005) Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot Cell 4:1942–1950. https://doi.org/10.1128/EC.4.11.1942-1950.2005
Schwede A, Kramer S, Carrington M (2012) How do trypanosomes change gene expression in response to the environment? Protoplasma 249:223–238. https://doi.org/10.1007/s00709-011-0282-5
Thelie A, Pascal G, Angulo L et al (2012) An oocyte-preferential histone mRNA stem-loop-binding protein like is expressed in several mammalian species. Mol Reprod Dev 79:380–391. https://doi.org/10.1002/mrd.22040
Urbaniak MD, Martin DMA, Ferguson MAJ (2013) Global quantitative SILAC phosphoproteomics reveals differential phosphorylation is widespread between the procyclic and bloodstream form lifecycle stages of Trypanosoma brucei. J Proteome Res 12:2233–2244. https://doi.org/10.1021/pr400086y
Vasquez J-J, Hon C-C, Vanselow JT et al (2014) Comparative ribosome profiling reveals extensive translational complexity in different Trypanosoma brucei life cycle stages. Nucleic Acids Res 42:3623–3637. https://doi.org/10.1093/nar/gkt1386
von Moeller H, Lerner R, Ricciardi A et al (2013) Structural and biochemical studies of SLIP1–SLBP identify DBP5 and eIF3g as SLIP1-binding proteins. Nucleic Acids Res 41:7960–7971. https://doi.org/10.1093/nar/gkt558
Walrad P, Paterou A, Acosta-Serrano A, Matthews KR (2009) Differential trypanosome surface coat regulation by a CCCH protein that co-associates with procyclin mRNA cis-elements. PLoS Pathog 5:e1000317. https://doi.org/10.1371/journal.ppat.1000317
Wang ZF, Ingledue TC, Dominski Z et al (1999) Two Xenopus proteins that bind the 3′ end of histone mRNA: implications for translational control of histone synthesis during oogenesis. Mol Cell Biol 19:835–845
Wurst M, Seliger B, Jha BA et al (2012) Expression of the RNA recognition motif protein RBP10 promotes a bloodstream-form transcript pattern in Trypanosoma brucei. Mol Microbiol 83:1048–1063. https://doi.org/10.1111/j.1365-2958.2012.07988.x
Yoffe Y, Zuberek J, Lerer A et al (2006) Binding specificities and potential roles of isoforms of eukaryotic initiation factor 4E in Leishmania. Eukaryot Cell 5:1969–1979. https://doi.org/10.1128/EC.00230-06
Yoffe Y, Léger M, Zinoviev A et al (2009) Evolutionary changes in the Leishmania eIF4F complex involve variations in the eIF4E–eIF4G interactions. Nucleic Acids Res 37:3243–3253. https://doi.org/10.1093/nar/gkp190
Zamudio JR, Mittra B, Chattopadhyay A et al (2009) Trypanosoma brucei spliced leader RNA maturation by the cap 1 2′-O-ribose methyltransferase and SLA1 H/ACA snoRNA pseudouridine synthase complex. Mol Cell Biol 29:1202–1211. https://doi.org/10.1128/MCB.01496-08
Zeiner GM, Foldynová S, Sturm NR et al (2004) SmD1 is required for spliced leader RNA biogenesis. Eukaryot Cell 3:241–244
Zhao X, McKillop-Smith S, Müller B (2004) The human histone gene expression regulator HBP/SLBP is required for histone and DNA synthesis, cell cycle progression and cell proliferation in mitotic cells. J Cell Sci 117:6043–6051. https://doi.org/10.1242/jcs.01523
Zheng L, Dominski Z, Yang X-C et al (2003) Phosphorylation of stem-loop binding protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol Cell Biol 23:1590–1601
Zinoviev A, Léger M, Wagner G, Shapira M (2011) A novel 4E-interacting protein in Leishmania is involved in stage-specific translation pathways. Nucleic Acids Res 39:8404–8415. https://doi.org/10.1093/nar/gkr555
Zinoviev A, Manor S, Shapira M (2012) Nutritional stress affects an atypical cap-binding protein in Leishmania. RNA Biol 9:1450–1460. https://doi.org/10.4161/rna.22709
Thompson MK, Gilbert WV (2017) mRNA length-sensing in eukaryotic translation: reconsidering the “closed loop” and its implications for translational control. Curr Genet 63:613–620. https://doi.org/10.1007/s00294-016-0674-3
Acknowledgements
The authors thank Dr. Mark Carrington, from University of Cambridge (UK), and Dr. Steve Kelly, from Department of Plant Sciences, University of Oxford (UK) for providing the SLBP protein sequences from Euglena gracilis. We also thank Dr. Cássia Docena from the Núcleo de Plataformas Tecnológicas (NPT), Instituto Aggeu Magalhães, Fiocruz-PE, for the assistance in obtaining Immunofluorescence images. The anti-BiP and anti-TY antibodies were kind gifts from Jay Bangs and Keith Gull, respectively. This work was supported by a UCLA Stein-Oppenheimer award (D. C.), the National Institutes of Health (Grant numbers AI056034, AI073806, TW009035 to D. C. and N. S., Grant number GM089778 to J. W.), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq-Brazil, Grant number 401282/2014-7) and Fundação Oswaldo Cruz-Fiocruz (Brazil). Funding was provided by Fogarty International Center (PAR-08-222).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
No potential conflicts of interest were disclosed.
Additional information
Communicated by M. Kupiec.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary figure 1. Multiple sequence alignment comparing the SLBP1 amino acid sequences from selected trypanosomatid species and the related
B. saltans ortholog. The RBD domain is highlighted by dashes with asterisks indicating conserved residues involved in mRNA stem-loop binding. Alignment indicates conserved regions of SLBP1 from B. saltans (BsSLBP1, GeneDB#: BSAL_11875; NCBI#: CUG87777.1), T. brucei (TbSLBP1, GeneDB#: Tb927.3.1910; NCBI#: XP_XP_843785.1), T. cruzi (TcSLBP1, GeneDB#: TcCLB.430061.9; NCBI#: XP_802584.1), L. major (LmSLBP1, GeneDB#: LmjF.25.1830; NCBI#: XP_001683942.1) and L. braziliensis (LbSLBP1, GeneDB#: LbrM.25.1390; NCBI# XP_001565643.1). Identical amino acids are indicated by black shading. Amino acids defined as similar, by the BLOSUM 62 Matrix, in >50% of the sequences are shaded gray Supplementary material 1 (EPS 2587 KB)
Supplementary figure 2. Multiple sequence alignment comparing the SLBP2 amino acid sequences from selected trypanosomatid species and the related
B. saltans ortholog. The RBD domain is highlighted by dashes with asterisks indicating conserved residues involved in mRNA stem-loop binding. The red box shows the conserved region found in SLBP2 orthologs only from B. saltans (BsSLBP2, GeneDB#: BSAL_85420; Pubmed#: CUG77401.1), T. brucei (TbSLBP2, GeneDB#: Tb927.03.870; Pubmed#: XP_843683.1), T. cruzi (TcSLBP2, GeneDB#: TcCLB.511867.150; Pubmed#: XP_814962.1), L. major (LmSLBP2, GeneDB#: LmjF.25.0920; Pubmed#: XP_001683846.1) and L. braziliensis (LbSLBP2, GeneDB#: LbrM.25.0800; Pubmed# XP_001565585.1). Identical amino acids are indicated by black shading. Amino acids defined as similar, by the BLOSUM 62 Matrix, in >60% of the sequences are shaded gray Supplementary material 2 (EPS 2440 KB)
Supplementary figure 3. Yeast two-hybrid assay confirms lack of interaction between TbEIF4E2 and the five
T. brucei EIF4G homologs. Interactions between TbEIF4E2 and TbEIF4G1 trough G5 homologs was assayed using the yeast two-hybrid assay in the presence of increasing amounts of 3AT, increasing the stringency of the assay. Positive controls were pGADT7-T and pGBKT7-53; the negative controls were the empty vectors Supplementary material 3 (EPS 1252 KB)
Supplementary figure 4. TbSLBP2-PTP construction
. (A) Schematic representation showing the TbSLBP2 cell line construction with integration of the PTP tag into the allele to generate the TbSLBP2wt/PTP cell line (same strategy was used for TbSLBP1wt/PTP cell line). (B) Western blot of extracts from the TbSLBP1wt/PTP and TbSLBP2wt/PTP cell lines using an antibody against the protein A domain of the PTP tag Supplementary material 4 (EPS 1408 KB)
Supplementary figure 5. mRNA levels of TbEIF4E2 and TbSLBP2
. Combined data from the transcriptome of T. brucei shows differential abundance over the cell cycle in procyclic cells for TbEIF4E2 and TbSLBP2 but not TbSLBP1 Supplementary material 5 (EPS 1050 KB)
Supplementary table 1.
. (XLSX 11 KB)
Supplementary table 2.
. (XLSX 17 KB)
Rights and permissions
About this article
Cite this article
Freire, E.R., Moura, D.M.N., Bezerra, M.J.R. et al. Trypanosoma brucei EIF4E2 cap-binding protein binds a homolog of the histone-mRNA stem-loop-binding protein. Curr Genet 64, 821–839 (2018). https://doi.org/10.1007/s00294-017-0795-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-017-0795-3