Abstract
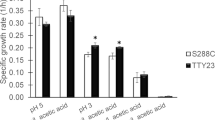
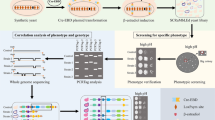
Adaptation by natural selection might improve the fitness of an organism and its probability to survive in unfavorable environmental conditions. Decoding the genetic basis of adaptive evolution is one of the great challenges to deal with. To this purpose, Saccharomyces cerevisiae has been largely investigated because of its short division time, excellent aneuploidy tolerance and the availability of the complete sequence of its genome with a thorough genome database. In the past, we developed a system, named bridge-induced translocation, to trigger specific, non-reciprocal translocations, exploiting the endogenous recombination system of budding yeast. This technique allows users to generate a heterogeneous population of cells with different aneuploidies and increased phenotypic variation. In this work, we demonstrate that ad hoc chromosomal translocations might induce adaptation, fostering selection of thermo-tolerant yeast strains with improved phenotypic fitness. This “yeast eugenomics” correlates with a shift to enhanced expression of genes involved in stress response, heat shock as well as carbohydrate metabolism. We propose that the bridge-induced translocation is a suitable approach to generate adapted, physiologically boosted strains for biotechnological applications.





Similar content being viewed by others
References
Anderson CA, Roberts S, Zhang H, Kelly CM, Kendall A, Lee C, Gerstenberger J, Koenig AB, Kabeche R, Gladfelter AS (2015) Ploidy variation in multinucleate cells changes under stress. Mol Biol Cell 26:1129–1140
Aragona M, Valente MT (2015) Genetic transformation of the tomato pathogen Pyrenochaeta lycopersici allowed gene knockout using a split-marker approach. Curr Genet 61:211–220
Arnak R, Altun B, Tosato V, Bruschi CV (2016) Multiple antibiotic resistance plasmids allow scalable PCR-mediated DNA manipulation and near-zero background cloning. Food Technol Biotechnol. doi:10.17113/ftb.54.03.16.4230
Auesukaree C, Koedrith P, Saenpayavai P, Asvarak T, Benjaphokee S, Sugiyama M, Kaneko Y, Harashima S, Boonchird C (2012) Characterization and gene expression profiles of thermotolerant S. cerevisiae isolates from Thai fruits. J Biosci Bioeng 114:144–149
Brion C, Ambroset C, Sanchez I, Legras JL, Blondin B (2013) Differential adaptation to multi-stressed conditions of wine fermentation revealed by variations in yeast regulatory networks. BMC Genom 14:681
Burke DT, Carle GF, Olson MV (1987) Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science 236:806–812
Camarasa C, Sanchez I, Brial P, Bigey F, Dequin S (2011) Phenotypic landscape of S. cerevisiae during wine fermentation: evidence for origin-dependent metabolic traits. PLoS One 6:e25147
Chan CY, Zhu J, Schiestl RH (2011) Effect of rad50 mutation on illegitimate recombination in S. cerevisiae. Mol Genet Genomics 285:471–484
Chang SL, Lai HY, Tung SY, Leu JY (2013) Dynamic large-scale chromosomal rearrangements fuel rapid adaptation in yeast populations. PLoS Genet 9:e1003232
Divol B, du Toit M, Duckitt E (2012) Surviving in the presence of sulphur dioxide: strategies developed by wine yeasts. Appl Microbiol Biotechnol 95:601–613
Dodgson SE, Kim S, Costanzo M, Baryshnikova A, Morse DL, Kaiser CA, Boone C, Amon A (2016) Chromosome specific and global effects of aneuploidy in S. cerevisiae. Genetics 202:1395–1409
Dudasova Z, Dudas A, Chovanec M (2004) Non-homologous end-joining factors of S. cerevisiae. FEMS Microbiol Rev 28:581–601
Gao J, Kan F, Wagnon JL, Storey AJ, Protacio RM, Davidson MK, Wahls WP (2014) Rapid, efficient and precise allele replacement in the fission yeast Schizosaccharomyces pombe. Curr Genet 60:109–119
Gibney PA, Lu C, Caudy AA, Hess DC, Botstein D (2013) Yeast metabolic and signaling genes are required for heat-shock survival and have little overlap with the heat-induced genes. Proc Natl Acad Sci USA 110:E4393–E4402
Goarin A, Silar P, Malagnac F (2015) Gene replacement in Penicillium roqueforti. Curr Genet 61:203–210
Kang GY, Kim EH, Lee HJ, Gil NY, Cha HJ, Lee YS (2015) Heat schock factor 1, an inhibitor of non-homologous end joining repair. Oncotarget 6:29712–29724
Kikukawa H, Sakuradani E, Nakatani M, Ando A, Okuda T, Sakamoto T, Ochiai M, Shimizu S, Ogawa J (2015) Gene targeting in the oil-producing fungus Mortierella alpina 1S-4 and construction of a strain producing a valuable polyunsaturated fatty acid. Curr Genet 61:579–589
Klinner U, Schäfer B (2004) Genetic aspects of targeted insertion mutagenesis in yeast. FEMS Microbiol Rev 28:201–223
Kraus E, Leung WY, Haber JE (2001) Break-induced replication: a review and an example in budding yeast. Proc Natl Acad Sci USA 98:8255–8262
Legras JL, Erny C, Jeune CL, Lollier M, Adolphe Y, Demuyter C, Delobel P, Blondin B, Karst F (2010) Activation of two different resistance mechanisms in S. cerevisiae upon exposure to octanoic and decanoic acids. Appl Environ Microbiol 76:7526–7535
Mjelle R, Hegre SA, Aas PA, Slupphaug G, Drabløs E, Saetrom P, Krokan HE (2015) Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair 30:53–67
Nakamura T, Yamamoto M, Saito K, Ando A, Shima J (2014) Identification of a gene, FMP21, whose expression levels are involved in thermotolerance in S. cerevisiae. AMB Express 4:67
Nikitin D, Tosato V, Zavec AB, Bruschi CV (2008) Cellular and molecular effects of non-reciprocal chromosome translocation in S. cerevisiae. Proc Natl Acad Sci USA 105:9703–9708
Nikitin D, Bruschi CV, Sims J, Breitenbach M, Rinnerthaler M, Tosato V (2014) Chromosome translocation may lead to PRK1-dependent anticancer drug resistance in yeast via endocytic actin network deregulation. Eur J Cell Biol 93:145–156
Randolph LF (1932) Some effects of high temperature on polyploidy and other variations in maize. Proc Natl Acad Sci USA 18:222–229
Riezman H (2004) Why do cells require heat shock proteins to survive heat stress? Cell Cycle 3:61–63
Rossi B, Noel P, Bruschi CV (2010) Different aneuploidies arise from the same bridge- induced chromosomal translocation event in S. cerevisiae. Genetics 186:775–790
Roukos V, Misteli T (2014) The biogenesis of chromosomal translocations. Nat Cell Biol 16:293–300
Satomura A, Katsuyama Y, Miura N, Kuroda K, Tomio A, Bamba T, Fukusaki E, Ueda M (2013) Acquisition of thermotolerant yeast S. cerevisiae by breeding via stepwise adaptation. Biotechnol Prog 29:1116–1123
Savitree L, Chutim AS, Wichien Y (2007) Production of fuel ethanol at high temperature from sugar cane juice by a newly isolated K. marxianus. Bioresour Technol 98:3367–3374
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C (T) method. Nat Protoc 3:1101–1108
Selmecki AM, Maruvka YE, Richmond PA, Guillet M, Shoresh N, Sorenson AL, De S, Kishony R, Michor F, Dowell R (2015) Polyploidy can drive rapid adaptation in yeast. Nature 519:349–352
Sims J, Bruschi CV, Bertin C, West N, Breitenbach M, Schroeder S, Eisenberg T, Rinnerthaler M, Raspor P, Tosato V (2016) High reactive oxygen species levels are detected at the end of the chronological life span of translocant yeast cells. Mol Genet Genom 291:423–435
Sridhar M, Kiran SN, Venkateswar RL (2002) Effect of UV radiation on thermotolerance, ethanol tolerance and osmotolerance of S. cerevisiae VS1 and VS3 strains. Bioresour Technol 83:199–202
Suhane T, Laskar S, Advani S, Roy N, Varunan S, Bhattacharyya D, Bhattacharyya S, Battacharyya MK (2015) Both the charged linker region and ATPase domain of Hsp90 are essential for Rad51-dependent DNA repair. Eukaryot Cell 14:64–77
Teste MA, Duquenne M, François JM, Parrou JL (2009) Validation of reference genes for quantitative expression analysis by real-time RT-PCR in S. cerevisiae. BMC Mol Biol 10:99
Tosato V, Bruschi CV (2015) Per aspera ad astra: when harmful chromosomal translocation become a plus value in genetic evolution. Lessons from S. cerevisiae. Microbial Cell 2:363–375
Tosato V, Waghmare SK, Bruschi CV (2005) Non-reciprocal chromosomal bridge- induced translocation (BIT) by targeted DNA integration in yeast. Chromosoma 114:15–27
Tosato V, Nicolini C, Bruschi CV (2009) DNA bridging of yeast chromosomes VIII leads to near-reciprocal translocation and loss of heterozygosity with minor cellular defects. Chromosoma 118:179–191
Tosato V, Sidari S, Bruschi CV (2013a) Bridge-induced chromosome translocation in yeast relies upon a Rad54/Rdh54-dependent, Pol32-independent pathway. PLoS One 8:e60926
Tosato V, Grüning NM, Breitenbach M, Arnak R, Ralser M, Bruschi CV (2013b) Genomic instability and Warburg effect: two yeast models for cancer cells. Front Oncol 2:212. doi:10.3389/fonc.2012.00212
Unnikrishnan I, Miller S, Meinke M, La Porte DC (2003) Multiple positive and negative elements involved in the regulation of expression of GSY1 in S. cerevisiae. J Biol Chem 278:26450–26457
Wach A, Brachat A, Pöhlmann R, Philippsen P (1994) New heterologous modules for classical or PCR-based gene disruptions in S. cerevisiae. Yeast 10:1793–1808
Wallace-Salinas V, Gorwa-Grauslund MF (2013) Adaptive evolution of an industrial strain of S. cerevisiae for combined tolerance to inhibitors and temperature. Biotechnol Biofuels 6:151
Wurtele H, Little KCE, Chartrand P (2003) Illegitimate DNA integration in mammalian cells. Gene Ther 10:1791–1799
Zang Q, Fu Y, Wang Y, Han J, Lv J, Wang S (2011) Improved ethanol production of a newly isolated thermo-tolerant S. cerevisiae strain after high-energy-pulse- electron beam. J Appl Microbiol 112:280–288
Zimmer A, Durand C, Loira N, Durrens P, Sherman DJ, Marullo P (2014) QTL dissection of lag phase in wine fermentation reveals a new translocation responsible for S. cerevisiae adaptation to sulfites. PLoS One 9:e86298
Acknowledgments
The authors wish to thank Jean-Luc Parou for having provided the plasmid pJL49 and Beatrice Rossi for having shared useful material and unpublished observations on SUSU translocants.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Funding
NW and VT were supported by Prosol SpA (I) and Crescendo Biologics Ltd (UK), respectively.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the Authors.
Additional information
Communicated by M. Kupiec.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tosato, V., Sims, J., West, N. et al. Post-translocational adaptation drives evolution through genetic selection and transcriptional shift in Saccharomyces cerevisiae . Curr Genet 63, 281–292 (2017). https://doi.org/10.1007/s00294-016-0635-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-016-0635-x