Abstract
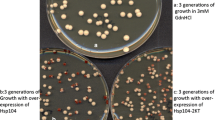
The yeast prion [PSI +] is a self-perpetuating aggregated isoform of the translation termination factor Sup35. [PSI +] propagation is promoted by moderate levels and antagonized by high levels of the chaperone Hsp104. In agreement with the model postulating that excess Hsp104 acts on [PSI +] by disaggregating prion polymers, we show that an increase in Sup35 levels, accompanied by an increase in size of prion aggregates, also partially protects [PSI +] from elimination by excess Hsp104. Despite retention of [PSI +], excess Hsp104 decreases toxicity of overproduced Sup35 in [PSI +] strains. A heritable variant of [PSI +], which has been isolated and is maintained only in the presence of increased levels of Hsp104, is characterized by an abnormally large aggregate size, and exhibits an altered response to overproduction of the Hsp70 chaperone Ssa1. These features resemble the previously described prion generated by a deletion derivative of Sup35, but are not associated with any sequence alteration and are controlled exclusively at the protein level. Our data provide a proof of the existence of conditionally stable prion variants maintained only at altered levels of Hsps, that could in principle be beneficial if the normal cellular function of a prion protein becomes detrimental to the cell in such conditions.



Similar content being viewed by others
References
Allen KD, Wegrzyn RD, Chernova TA, Müller S, Newnam GP, Winslett PA, Wittich KB, Wilkinson KD, Chernoff YO (2005) Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 169:1227–1242
Bailleul-Winslett PA, Newnam GP, Wegrzyn RD, Chernoff YO (2000) An antiprion effect of the anticytoskeletal drug latrunculin A in yeast. Gene Exp 9:145–156
Borchsenius AS, Wegrzyn RD, Newnam GP, Inge-Vechtomov SG, Chernoff YO (2001) Yeast prion protein derivative defective in aggregate shearing and production of new seeds. EMBO J 20:6683–6691
Chernoff YO (2004a) Cellular control of prion formation and propagation in yeast. In: Telling G (ed) Prions and prion diseases: current perspectives. Horizon Scientific Press, Norfolk, pp 257–303
Chernoff YO (2004b) Amyloidogenic domains, prions and structural inheritance: rudiments of early life or recent acquisition? Curr Opin Chem Biol 8:665–671
Chernoff YO, Inge-Vechtomov SG, Derkach IL, Ptyushkina MV, Tarunina OV, Dagkesamanskaya AR, Ter-Avanesyan MD (1992) Dosage-dependent translational suppression in yeast Saccharomyces cerevisiae. Yeast 8:489–499
Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi +]. Science 268:880–884
Chernoff YO, Newnam GP, Kumar J, Allen K, Zink AD (1999) Evidence for a “protein mutator” in yeast: role of the Hsp70-related chaperone Ssb in formation, stability and toxicity of the [PSI] prion. Mol Cell Biol 19:8103–8112
Chernoff YO, Galkin AP, Lewitin E, Chernova TA, Newnam GP, Belenkiy SM (2000) Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol Microbiol 35:865–876
Chernoff YO, Uptain SM, Lindquist SL (2002) Analysis of prion factors in yeast. Meth Enzymol 351:499–538
Cox B, Ness F, Tuite M (2003) Analysis of the generation and segregation of propagons: entities that propagate the [PSI +] prion in yeast. Genetics 165:23–33
Dagkesamanskaya AR, Ter-Avanesyan MD (1991) Interaction of the yeast omnipotent suppressors SUP1 (SUP45) and SUP2 (SUP35) with non-mendelian factors. Genetics 128:513–520
Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW (1996) Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144:1375–1386
Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW (1997) Genetic and environmental factors affecting the de novo appearance of the [PSI +] prion in Saccharomyces cerevisiae. Genetics 147:507–519
Gari E, Piedrafita L, Aldea M, Herrero E (1997) A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13:837–848
Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV (2003) Yeast [PSI +] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem 278:49636–49643
Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB (2005) Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci USA 102:10575–10580
Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO (1999) Antagonistic interactions between yeast chaperones Hsp104 and Hsp70. Mol Cell Biol 19:1325–1333
Osherovich LZ, Cox BS, Tuite MF, Weissman JS (2004) Dissection and design of yeast prions. PLoS Biol 2(4):E86. DOI 10.1371/journal.pbio.0020086
Patino MM, Liu JJ, Glover JR, Lindquist S (1996) Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273:622–626
Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD (1996) Propagation of the yeast prion-like [psi +] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J 15:3127–3134
Resende CG, Outeiro TF, Sands L, Lindquist S, Tuite MF (2003) Prion protein gene polymorphisms in Saccharomyces cerevisiae. Mol Microbiol 49:1005–1017
Sanchez Y, Taulien J, Borkovich KA, Lindquist S (1992) Hsp104 is required for tolerance to many forms of stress. EMBO J 11:2357–2364
Sherman F (2002) Getting started with yeast. Meth Enzymol 350:3–41
True HL, Lindquist SL (2000) A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407:477–483
True HL, Berlin I, Lindquist SL (2004) Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature 431:184–187
Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO (2001) Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol 21:4656–4669
Weissmann C (2004) The state of the prion. Nat Rev Microbiol 2:861–871
Zhou P, Derkatch IL, Liebman SW (2001) The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI +] and [PIN +]. Mol Microbiol 39:37–46
Acknowledgements
We thank K. Gokhale and E. Lewitin for help in the strain and plasmid constructions, and D. Bedwell, S. Lindquist, and J. Weissman for plasmids and antibodies. This work was supported by grants RB1-2336-ST02 to S. G. I.-V. and Y. O. C.; and ST-012-02 (YI-B-12-06) to A. S. B. from US Civilian Research and Development Foundation; R01GM58763 to Y. O. C. from National Institutes of Health; and 03-04-49335 to A. S. B. from Russian Foundation for Basic Research.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Liebman
A. S. Borchsenius and S. Müller contributed equally to this study.
Rights and permissions
About this article
Cite this article
Borchsenius, A.S., Müller, S., Newnam, G.P. et al. Prion variant maintained only at high levels of the Hsp104 disaggregase. Curr Genet 49, 21–29 (2006). https://doi.org/10.1007/s00294-005-0035-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-005-0035-0