Abstract
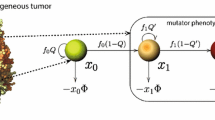
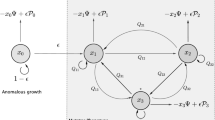
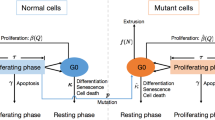
The dynamics of heterogeneous tumor cell populations competing with healthy cells is an important topic in cancer research with deep implications in biomedicine. Multitude of theoretical and computational models have addressed this issue, especially focusing on the nature of the transitions governing tumor clearance as some relevant model parameters are tuned. In this contribution, we analyze a mathematical model of unstable tumor progression using the quasispecies framework. Our aim is to define a minimal model incorporating the dynamics of competition between healthy cells and a heterogeneous population of cancer cell phenotypes involving changes in replication-related genes (i.e., proto-oncogenes and tumor suppressor genes), in genes responsible for genomic stability, and in house-keeping genes. Such mutations or loss of genes result into different phenotypes with increased proliferation rates and/or increased genomic instabilities. Despite bifurcations in the classical deterministic quasispecies model are typically given by smooth, continuous shifts (i.e., transcritical bifurcations), we here identify a novel type of bifurcation causing an abrupt transition to tumor extinction. Such a bifurcation, named as trans-heteroclinic, is characterized by the exchange of stability between two distant fixed points (that do not collide) involving tumor persistence and tumor clearance. The increase of mutation and/or the decrease of the replication rate of tumor cells involves this catastrophic shift of tumor cell populations. The transient times near bifurcation thresholds are also characterized, showing a power law dependence of exponent \(-1\) of the transients as mutation is changed near the bifurcation value. These results are discussed in the context of targeted cancer therapy as a possible therapeutic strategy to force a catastrophic shift by simultaneously delivering mutagenic and cytotoxic drugs inside tumor cells.





Similar content being viewed by others
References
Adams J, Bellomo N (1997) A survey of models for tumor-immune system dynamics. Birkhäuser
Anderson GR, Stoler DL, Brenner BM (2001) Cancer as an evolutionary consequence of a destabilized genome. Bioessays 23:103746
Bi P, Ruan S, Zhang X (2014) Periodic and chaotic oscillations in a tumor and immune system interaction model with three delays. Chaos 24:023101
Bielas JH, Loeb KR, Rubin BP, True LD et al (2006) Human cancers express a mutator phenotype. Proc Natl Acad Sci USA 103:18238–18242
Bjedov I, Tenaillon O, Gérard B, Souza V et al (2003) Stress-induced mutagenesis in bacteria. Science 300:1404–1409
Bowman P, Jonkers J (2012) The effects of deregulated DNA damage signaling on cancer chemotherapy response and resistance. Nat Rev Cancer 12:587–598
Bull JJ, Meyers LA, Lachmann M (2005) Quasispecies made simple. PLOS Comput Biol 1(6):e61
Bull JJ, Sanjuán R, Wilke CO (2007) Theory of lethal mutagenesis for viruses. J Virol 81(6):2930–2939
Cahill DP, Kinzler KW, Vogelstein B, Lengauer C (1999) Genetic instabilities and Darwinian selection in tumors. Trends Genet 15:M57–61
Castillo V, Lázaro JT, Sardanyés J (2016) Dynamics and bifurcations in a simple quasispecies model of tumorigenesis. Comp Appl Math (in press)
Chari RV (2008) Targeted cancer therapy: conferring specificity to cytotoxic drugs. Acc Chem Res 41(1):98–107
Domingo E, Sheldon J, Perales C (2012) Viral quasispecies evolution. Microbiol Mol Biol 7:159–216
Eigen M (1971) Self-organization of matter and the evolution of biological macromolecules. Naturwiss 58:465–523
Eigen M, McCaskill J, Schuster P (1989) The molecular quasi-species. Adv Chem Phys 75:149–263
Eisenberg E, Levanon EY (2003) Housekeeping genes are compact. Trends Genet 19:362–366
Feig DI, Loeb LA (1992) Mechanisms of mutation by oxidative DNA damage: reduced fidelity of mammalian DNA polymerase \(\beta \). Biochemistry 32:4466–4473
Fornari C, Balbo G, Halawani SM, Ba-Rukab O, Ahmad AR, Calogero RA et al (2015) A versatile mathematical work-flow to explore how cancer stem cell fate influences tumor progression. BMC Syst Biol 9:1–20
Fox EJ, Loeb LA (2010) Lethal mutagenesis: targeting the mutator phenotype in cancer. Sem Cancer Biol 20:353–359
Franz S, Peliti L (1997) Error threshold in simple landscapes. J Phys A Math Gen 30:4481–4487
Garay RP, Lefever R (1978) A kinetic approach of the immunology of cancer: stationary states properties of effector-target cell reactions. J Theor Biol 73:417–438
Gatenby RA, Frieden BR (2002) Application of information theory and extreme physical information to carcinogenesis. Cancer Res 62:3675–3684
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100:57–70
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674
Helleday H, Petermann E, Lundin C, Hodgson B et al (2008) DNA repair pathways as targets for cancer therapy. Nat Rev Cancer 8:193–200
Hoeijmakers JH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411:366–374
Itik M, Banks SP (2010) Chaos in a three-dimensional cancer model. Int J Bifurc Chaos 20:71–79
Kirschner D, Oanetta JC (1998) Modeling immunotherapy of the tumor-immune interaction. J Math Biol 37:235–252
Komarova NL, Wodarz D (2003) Evolutionary dynamics of mutator phenotypes in cancer: implications and chemotherapy. Cancer Res 63:6635–6642
Kuznetsov VA, Makalkin IA, Taylor MA, Perelson AS (1994) Nonlinear dynamics of immunogenic tumors: parameter estimation and global bifurcation analysis. Bull Math Biol 56:295–321
Lengauer C, Kinzler KW, Vogelstein B (1998) Genetic instabilities in human cancers. Nature 396:643–649
Loeb LA, Essigman JM, Kazazi F, Zhang J, Rose KD, Mullins JI (1999) Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc Natl Acad Sci USA 96:1492–1497
Loeb LA (2001) A mutator phenotype in cancer. Cancer Res 61:3230–3239
Loeb LA (2011) Human cancers express the mutator phenotypes: origin, consequences and targeting. Nature 11:450–457
Malgorzata E, Jonkers J (2011) Studying therapy response and resistance in mouse models for BRCA-1deficient breast cancer. J Mammary Gland Biol Neoplasia 16:41–50
Merlo LMF, Pepper JW, Reid BJ, Maley CC (2006) Cancer as an evolutionary and ecological process. Nat Rev Cancer 6:924–935
Oliver A, Canton R, Campo P, Baquero F et al (2000) High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1254
O’Neil NJ, van Pel DM, Hieter P (2013) Synthetic lethality and cancer: cohesin and PARP at the replication fork. Trends Genet 8:e1002574
Rayner E, van Gool IC, Palles C, Kearsey SE et al (2016) A panoply of errors: polymerase proofreading domain mutations in cancer. Nat Rev 16:71–81
Sardanyés J, Simó C, Martínez R, Solé RV, Elena SF (2014) Variability in mutational fitness effects prevents full lethal transitions in large quasispecies populations. Sci Rep 4:4625
Sardanyés J, Elena SF (2010) Error threshold in RNA quasispecies models with complementation. J Theor Biol 265(3):278-86
Sniegowski PD, Gerrish PJ, Lenski RE (1997) Evolution of high mutation rates in experimental populations of E. coli. Nature 387:703–705
Solé RV (2003) Phase transitions in unstable cancer cell populations. Eur J Phys B 35:117–124
Solé RV (2012) Phase transitions in cancer. In: d’Onofrio A et al. (eds.) New challenges for cancer systems biomedicine. Springer, pp 35–51
Solé RV, Sardanyés J, Díez J, Mas A (2006) Information catastrophe in RNA viruses through replication thresholds. J Theor Biol 240(3):353–359
Solé RV, Rodríguez-Caso C, Deisboeck TS, Saldaña J (2008) Cancer stem cells as the engine of unstable tumor progression. J Theor Biol 253:629–637
Solé RV, Valverde S, Rodriguez-Caso C, Sardanyés J (2014) Can a minimal replicating construct be identified as the embodiment of cancer? Bioessays 36:503–512
Solé RV, Deisboeck TS (2004) An error catastrophe in cancer? J Theor Biol 228:47–54
Sun W, Jiang T, Lu Y, Reiff M, Mo R, Gu Z (2014) Cocoon-like self-degradable DNA nano clew for anticancer drug delivery. J Am Chem Soc 136:14722–14725
Vogelstein B, Kinzler KW (2004) Cancer genes and the pathways they control. Nat Med 10:789–799
Wilke CO, Ronnewinkel C, Martinetz T (2001) Dynamic fitness landscapes in molecular evolution. Phys Rep 349:395–446
Wylie CS, Shakhnovic EL (2012) Mutation induced extinction in finite populations: lethal mutagenesis and lethal isolation. PLOS Comput Biol 8:e1002609
Yue QX, Liu X, Guo DA (2010) Microtubule-binding natural products for cancer therapy. Planta Medica 76(11):1037–1043
Acknowledgments
We thank the members of the Complex Systems Lab for their helpful comments, as well as the members of the Institute of Evolutionary Biology (IBE) for their critics and comments. The authors acknowledge the computing facilities of the Dynamical Systems Group (Universitat de Barcelona). This article was partially funded by the Botín Foundation (JS, RVS), by the Spanish grant FIS2012-39288 (RVS), by the Santa Fe Institute (RVS), by the Spanish Secretaria de Estado de Investigación, Desarrollo e Innovación grants MTM2013-41168-P (CS, RM), and by grant 2014-SGR-1145 from the Catalan government (CS, RM). This work has also counted with the support of Secretaria d’Universitats i Recerca del Departament d’Economia i Coneixement de la Generalitat de Catalunya.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sardanyés, J., Martínez, R., Simó, C. et al. Abrupt transitions to tumor extinction: a phenotypic quasispecies model. J. Math. Biol. 74, 1589–1609 (2017). https://doi.org/10.1007/s00285-016-1062-9
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-016-1062-9
Keywords
- Applied mathematics
- Bifurcations
- Cancer evolution
- Dynamical systems
- Genomic instability
- Phenotypic model
- Quasispecies