Abstract
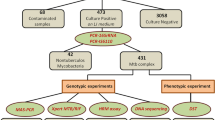
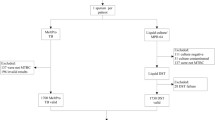
Multidrug-resistant tuberculosis (MDR-TB) requires treatment with fluoroquinolone (FLQ) drugs, however, the excessive use of FLQ has led to the rise of extensively drug-resistant TB. In 2019, ~ 20% of total MDR-TB cases were estimated to be resistant to FLQ drugs. In the present study, we developed and evaluated the utility of high-resolution melt curve analysis (HRM) for the rapid detection of FLQ-resistant Mycobacterium tuberculosis for the first time directly from sputum samples. A reference plasmid library was generated for the most frequently observed mutations of gyrA gene and was used to discriminate between mutant and wild-type samples in the FLQ-HRM assay. The developed assay was evaluated on n = 25 MDR M. tuberculosis clinical isolates followed by validation on archived sputum DNA (n = 88) using DNA sequencing as a gold standard. The FLQ-HRM assay showed a 100% sensitivity [95% Confidence Interval (CI): 71.5 to 100] and specificity (95% CI: 39.7 to 100) in smear-positive category, and a sensitivity of 88.9% (95% CI: 77.3 to 95.8) with 84.2% (95% CI: 60.4 to 96.6) specificity in smear-negative category. The assay showed a high level of concordance of ~ 90% (κ = 0.74) with DNA sequencing, however, we were limited by the absence of phenotypic drug susceptibility testing data. In conclusion, HRM is a rapid, cost-effective (INR 150/USD 1.83) and closed-tube method for direct detection of FLQ resistance in sputum samples including direct smear-negative samples.


Similar content being viewed by others
Data Availability
The data used or analysed during the present study are available from the corresponding author on reasonable request.
Code Availability
Not applicable.
References
WHO (2022) Global Tuberculosis Report.
WHO (2022) WHO consolidated guidelines on tuberculosis, Module 4: Treatment- Drug-resistant tuberculosis treatment: 2022 update.
WHO (2020) Meeting report of the WHO expert consultation on the definition of extensively drug-resistant tuberculosis.
Jabeen K, Shakoor S, Hasan R (2015) Fluoroquinolone-resistant tuberculosis: implications in settings with weak healthcare systems. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases 32:118–123. https://doi.org/10.1016/j.ijid.2015.01.006
WHO (2020) Global Tuberculosis Report.
WHO (2021) Information sheet: Practical considerations for implementation of the Cepheid Xpert MTB/XDR test.
WHO (2016) The use of molecular line probe assays for the detection of resistance to second-line anti-tuberculosis drugs: Policy Guidance.
(TAG) TAG (2022) Tuberculosis Diagnostics: Pipeline Report.
Anthwal D, Gupta RK, Bhalla M, Bhatnagar S, Tyagi JS, Haldar S (2017) Direct detection of rifampin and isoniazid resistance in sputum samples from tuberculosis patients by high-resolution melt curve analysis. J Clin Microbiol 55(6):1755–1766. https://doi.org/10.1128/JCM.02104-16
Keikha M, Karbalaei M (2021) High resolution melting assay as a reliable method for diagnosing drug-resistant TB cases: a systematic review and meta-analysis. BMC Infect Dis 21(1):989. https://doi.org/10.1186/s12879-021-06708-1
Micheni LN, Kassaza K, Kinyi H, Ntulume I, Bazira J (2021) Rifampicin and isoniazid drug resistance among patients diagnosed with pulmonary tuberculosis in southwestern Uganda. PLoS ONE 16(10):e0259221. https://doi.org/10.1371/journal.pone.0259221
Sharma K, Modi M, Kaur H, Sharma A, Ray P, Varma S (2015) rpoB gene high-resolution melt curve analysis: a rapid approach for diagnosis and screening of drug resistance in tuberculous meningitis. Diagn Microbiol Infect Dis 83(2):144–149. https://doi.org/10.1016/j.diagmicrobio.2015.06.010
Sharma K, Sharma M, Singh S, Modi M, Sharma A, Ray P, Varma S (2017) Real-time PCR followed by high-resolution melting curve analysis: a rapid and pragmatic approach for screening of multidrug-resistant extrapulmonary tuberculosis. Tuberculosis (Edinb) 106:56–61. https://doi.org/10.1016/j.tube.2017.07.002
Haldar S, Chakravorty S, Bhalla M, De Majumdar S, Tyagi JS (2007) Simplified detection of Mycobacterium tuberculosis in sputum using smear microscopy and PCR with molecular beacons. J Med Microbiol 56(Pt 10):1356–1362. https://doi.org/10.1099/jmm.0.47265-0
WHO (2021) WHO consolidated guidelines on tuberculosis, Module 3: Diagnosis- Rapid diagnostics for tuberculosis detection0- 2021 update.
WHO (2021) Update on the use of nucleic acid amplification tests to detect TB and drug-resistant TB: rapid communication.
Anthwal D, Gupta RK, Singhal R, Bhalla M, Verma AK, Khayyam KU, Myneedu VP, Sarin R, Gupta A, Gupta NK, Singh M, Sivaswami Tyagi J, Haldar S (2021) Compatibility of a novel filter paper-based bio-safe sputum transport kit with line probe assay for diagnosing drug-resistant tuberculosis: a single-site evaluation study. ERJ Open Res. https://doi.org/10.1183/23120541.00137-2021
Zimenkov DV, Antonova OV, Kuz’min AV, Isaeva YD, Krylova LY, Popov SA, Zasedatelev AS, Mikhailovich VM, Gryadunov DA (2013) Detection of second-line drug resistance in Mycobacterium tuberculosis using oligonucleotide microarrays. BMC Infect Dis 13:240. https://doi.org/10.1186/1471-2334-13-240
Chawla K, Kumar A, Shenoy VP, Chakrabarty S, Satyamoorthy K (2018) Genotypic detection of fluoroquinolone resistance in drug-resistant Mycobacterium tuberculosis at a tertiary care centre in south Coastal Karnataka, India. Journal of global antimicrobial resistance 13:250–253. https://doi.org/10.1016/j.jgar.2018.01.023
Pillay S, Steingart KR, Davies GR, Chaplin M, De Vos M, Schumacher SG, Warren R, Theron G (2022) Xpert MTB/XDR for detection of pulmonary tuberculosis and resistance to isoniazid, fluoroquinolones, ethionamide, and amikacin. Cochrane Database Syst Rev 5(5):CD014841. https://doi.org/10.1002/14651858.CD014841.pub2
Singh P, Jain A, Dixit P, Prakash S, Jaiswal I, Venkatesh V, Singh M (2015) Prevalence of gyrA and B gene mutations in fluoroquinolone-resistant and -sensitive clinical isolates of Mycobacterium tuberculosis and their relationship with MIC of ofloxacin. J Antibiot 68(1):63–66. https://doi.org/10.1038/ja.2014.95
Singhal R, Reynolds PR, Marola JL, Epperson LE, Arora J, Sarin R, Myneedu VP, Strong M, Salfinger M (2016) Sequence analysis of fluoroquinolone resistance-associated genes gyrA and gyrB in clinical Mycobacterium tuberculosis isolates from patients suspected of having multidrug-resistant tuberculosis in New Delhi. India J Clin Microbiology 54(9):2298–2305. https://doi.org/10.1128/JCM.00670-16
Potdar PT, P, (2013) Development of sequence based molecular diagnostic test to evaluate MDR and XDR in M tuberculosis Patients from Western India. Am J Infect Dis Microb 1(3):50–58
Chen X, Kong F, Wang Q, Li C, Zhang J, Gilbert GL (2011) Rapid detection of isoniazid, rifampin, and ofloxacin resistance in Mycobacterium tuberculosis clinical isolates using high-resolution melting analysis. J Clin Microbiol 49(10):3450–3457. https://doi.org/10.1128/JCM.01068-11
Lee AS, Ong DC, Wong JC, Siu GK, Yam WC (2012) High-resolution melting analysis for the rapid detection of fluoroquinolone and streptomycin resistance in Mycobacterium tuberculosis. PLoS ONE 7(2):e31934. https://doi.org/10.1371/journal.pone.0031934
Pholwat S, Liu J, Stroup S, Gratz J, Banu S, Rahman SM, Ferdous SS, Foongladda S, Boonlert D, Ogarkov O, Zhdanova S, Kibiki G, Heysell S, Houpt E (2015) Integrated microfluidic card with TaqMan probes and high-resolution melt analysis to detect tuberculosis drug resistance mutations across 10 genes. mBio 6(2):e02273. https://doi.org/10.1128/mBio.02273-14
Sirous M, Khosravi AD, Tabandeh MR, Salmanzadeh S, Ahmadkhosravi N, Amini S (2018) Molecular detection of rifampin, isoniazid, and ofloxacin resistance in Iranian isolates of Mycobacterium tuberculosis by high-resolution melting analysis. Infection and drug resistance 11:1819–1829. https://doi.org/10.2147/IDR.S178831
Silva JL, Leite GG, Bastos GM, Lucas BC, Shinohara DK, Takinami JS, Miyata M, Fajardo CM, Luchessi AD, Leite CQ, Cardoso RF, Hirata RD, Hirata MH (2013) Plasmid-based controls to detect rpoB mutations in Mycobacterium tuberculosis by quantitative polymerase chain reaction-high-resolution melting. Mem Inst Oswaldo Cruz 108(1):106–109. https://doi.org/10.1590/s0074-02762013000100017
Bentaleb EM, El Messaoudi MD, Abid M, Messaoudi M, Yetisen AK, Sefrioui H, Amzazi S, Ait Benhassou H (2017) Plasmid-based high-resolution melting analysis for accurate detection of rpoB mutations in Mycobacterium tuberculosis isolates from Moroccan patients. BMC Infect Dis 17(1):548. https://doi.org/10.1186/s12879-017-2666-4
WHO (2016) Tuberculosis Diagnostics: Molecular Line-Probe Assay for the detection of resistance to Second-Line Anti-TB drugs (SL-LPA).
Acknowledgements
Not applicable.
Funding
The work was supported by the intramural grant (71/2-Edu-16/4465) from PGIMER, Chandigarh and extramural grant from Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India (BT/PR20814/MED/29/1214/2017).
Author information
Authors and Affiliations
Contributions
Conceptualization: SH; Methodology: RKG, DA; Formal analysis: RKG, DA; Investigation and data curation RKG, DA, MB; Writing—original draft preparation: RKG, DA; Writing—review and editing: MB, JST, SC, SH; Funding acquisition: SH; Resources: SH; Supervision: SC, JST, SH.
Corresponding author
Ethics declarations
Conflicts of interest
We declare no conflict of interest.
Ethics Approval
An ethical approval was obtained from the Institutional Ethics Committee of the Postgraduate Institute of Medical Education and Research (PGIMER, INT/IEC/2018/1456), Chandigarh for the use of archived DNA samples.
Consent to Participate
Not applicable.
Consent to Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gupta, R.K., Anthwal, D., Bhalla, M. et al. Direct Detection of Fluoroquinolone Resistance in Sputum Samples from Tuberculosis Patients by High Resolution Melt Curve Analysis. Curr Microbiol 81, 27 (2024). https://doi.org/10.1007/s00284-023-03519-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03519-2