Abstract
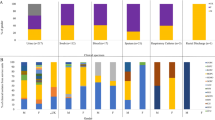
Klebsiella pneumoniae is a major drug-resistant human pathogen accountable for a wide range of infections. In this cross-sectional study, we aimed to determine the phenotypic and genotypic features of β-lactamase-producing K. pneumoniae clinical isolates from Alexandria, Egypt. A total of 50 nonduplicated clinical isolates of K. pneumoniae were obtained from various specimens. They were identified biochemically and by biotyping using mass spectrometry. For molecular characterization, plasmid profile analysis was performed. Screening for extended spectrum β-lactamases (ESBLs), carbapenemases and AmpC production was carried out phenotypically and genotypically. Correlation analysis was performed to assess the relationship between phenotype, genotype and resistance patterns among the studied isolates. The dendrogram demonstrated 38 distinct plasmid profiles among 62% of our isolates. According to antimicrobial susceptibility testing, 90% of isolates were multi/extensive-drug resistant. Nineteen out of 50 (38%) were resistant to cefoxitin, while only 10 (20%) were resistant to imipenem. All isolates were susceptible to colistin. Phenotypically, ESBL producers (78%) were the most common, followed by carbapenemase producers (24%). Genotypically, the most common ESBL gene was blaSHV (90%), followed by blaCTX-Mu (74%), while the most common carbapenemase genes were blaNDM (56%) and blaOXA-48 (54%). No blaKPC or blaIMP were detected. Plasmid-mediated AmpC resistance was confirmed in only two out of 19 cefoxitin-resistant isolates. Both the blaNDM and blaOXA.48 genes were significantly positive correlated (rho = 0.56, p = 0.004). Absence of blaKPC among carbapenem resistant K. pneumoniae isolates in Alexandria, Egypt. AmpC production is not the main factor behind the resistance to cefoxitin among our isolates.



Similar content being viewed by others
Data Availability
The authors confirm that the data supporting the results and findings of this study are available within the manuscript.
Code Availability
Not applicable.
References
Diekema DJ, Hsueh PR, Mendes RE, Pfaller MA, Rolston KV, Sader HS et al (2019) The Microbiology of bloodstream infection: 20-Year trends from the SENTRY antimicrobial surveillance program. Antimicrob Agents Chemother. https://doi.org/10.1128/aac.00355-19
Kobayashi K, Yamamoto S, Takahashi S, Ishikawa K, Yasuda M, Wada K et al (2020) The third national Japanese antimicrobial susceptibility pattern surveillance program: bacterial isolates from complicated urinary tract infection patients. J Infect Chemother 26(5):418–428. https://doi.org/10.1016/j.jiac.2020.01.004
Alfouzan W, Dhar R, Abdo NM, Alali WQ, Rabaan AA (2021) Epidemiology and microbiological profile of common healthcare associated infections among patients in the intensive care unit of a general hospital in kuwait: a retrospective observational study. J Epidemiol Glob Health 11(3):302–309. https://doi.org/10.2991/jegh.k.210524.001
Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL et al (2018) Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18(3):318–327. https://doi.org/10.1016/s1473-3099(17)30753-3
Hamam S, Sakr A, Zahran W, El Kholy R, Kasemy Z, Ibrahem R et al (2021) Health care-associated infections at an Egyptian tertiary care hospital: a 2-year prospective study. Menoufia Med J 34(2):514–520. https://doi.org/10.4103/mmj.mmj_455_20
Hassan R, El-Gilany AH, Abd Elaal AM, El-Mashad N, Azim DA (2020) An overview of healthcare-associated infections in a tertiary care hospital in Egypt. Infect Prev Pract. 2(3):100059. https://doi.org/10.1016/j.infpip.2020.100059
El-Kholy AA, Girgis SA, Shetta MAF, Abdel-Hamid DH, Elmanakhly AR (2020) Molecular characterization of multidrug-resistant gram-negative pathogens in three tertiary hospitals in Cairo. Egypt Eur J Clin Microbiol Infect Dis 39(5):987–992. https://doi.org/10.1007/s10096-020-03812-z
Lai CC, Yu WL (2021) Klebsiella pneumoniae harboring carbapenemase genes in Taiwan: its evolution over 20 years, 1998–2019. Int J Antimicrob Agents. 58(1):106354. https://doi.org/10.1016/j.ijantimicag.2021.106354
Bush K, Jacoby GA (2010) Updated functional classification of beta-lactamases. Antimicrob Agents Chemother 54(3):969–976. https://doi.org/10.1128/aac.01009-09
Ambler RP (1980) The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci 289(1036):321–331. https://doi.org/10.1098/rstb.1980.0049
Branka B, Sanda S (2018) Carbapenemases. In: Madhusmita M (ed) Growing and Handling of Bacterial Cultures. Rijeka, IntechOpen, p 3
Ma Y, Bao C, Liu J, Hao X, Cao J, Ye L et al (2018) Microbiological characterization of Klebsiella pneumoniae isolates causing bloodstream infections from five tertiary hospitals in Beijing. China J Glob Antimicrob Resist 12:162–166. https://doi.org/10.1016/j.jgar.2017.10.002
Peirano G, Sang JH, Pitondo-Silva A, Laupland KB, Pitout JD (2012) Molecular epidemiology of extended-spectrum-β-lactamase-producing Klebsiella pneumoniae over a 10 year period in Calgary. Canada J Antimicrob Chemother 67(5):1114–1120. https://doi.org/10.1093/jac/dks026
Hammoudi Halat D, Ayoub MC (2020) The current burden of carbapenemases: review of significant properties and dissemination among gram-negative bacteria. Antibiotics 9(4):186
Abdelaziz MO, Bonura C, Aleo A, Fasciana T, Mammina C (2013) NDM-1- and OXA-163-producing Klebsiella pneumoniae isolates in Cairo, Egypt, 2012. J Glob Antimicrob Resist 1(4):213–215. https://doi.org/10.1016/j.jgar.2013.06.003
Helal SF, El-Rachidi NG, AbdulRahman EM, Hassan DM (2014) The presence of blaKPC-mediated resistance in Enterobacteriaceae in Cairo University hospitals in Egypt and its correlation with in vitro carbapenem susceptibility. J Chemother 26(2):125–128. https://doi.org/10.1179/1973947813y.0000000099
Poirel L, Abdelaziz MO, Bernabeu S, Nordmann P (2013) Occurrence of OXA-48 and VIM-1 carbapenemase-producing Enterobacteriaceae in Egypt. Int J Antimicrob Agents 41(1):90–91. https://doi.org/10.1016/j.ijantimicag.2012.08.015
Altayb HN, Elbadawi HS, Alzahrani FA, Baothman O, Kazmi I, Nadeem MS et al (2022) Co-occurrence of β-lactam and aminoglycoside resistance determinants among clinical and environmental isolates of Klebsiella pneumoniae and Escherichia coli: a genomic approach. Pharmaceuticals (Basel, Switzerland). https://doi.org/10.3390/ph15081011
Salah FD, Soubeiga ST, Ouattara AK, Sadji AY, Metuor-Dabire A, Obiri-Yeboah D et al (2019) Distribution of quinolone resistance gene (qnr) in ESBL-producing Escherichia coli and Klebsiella spp. in Lomé Togo. Antimicrob Resist Infect Control 8:104. https://doi.org/10.1186/s13756-019-0552-0
Wassef M, Behiry I, Younan M, El Guindy N, Mostafa S, Abada E (2014) Genotypic identification of AmpC β-lactamases production in gram-negative Bacilli Isolates. Jundishapur J Microbiol 7(1):e8556. https://doi.org/10.5812/jjm.8556
Jacoby GA (2009) AmpC beta-lactamases. Clin Microbiol Rev 22(1):161–182. https://doi.org/10.1128/cmr.00036-08
Tille P. (2015) Bailey & Scott's diagnostic microbiology-E-Book: Elsevier Health Sciences
Singhal N, Kumar M, Kanaujia PK, Virdi JS (2015) MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol 6:791. https://doi.org/10.3389/fmicb.2015.00791
Perrier X, Jacquemoud-Collet, J.P. DARwin (2006) software [Available from: http://darwin.cirad.fr/.
Clinical and Laboratory Standards Institute (CLSI). (2020) Performance standards for antimicrobial susceptibility testing, 30th informational supplement M100-S30
Drieux L, Brossier F, Sougakoff W, Jarlier V (2008) Phenotypic detection of extended-spectrum β-lactamase production in Enterobacteriaceae: review and bench guide. Clin Microbiol Infect 14:90–103. https://doi.org/10.1111/j.1469-0691.2007.01846.x
Jarlier V, Nicolas MH, Fournier G, Philippon A (1988) Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis 10(4):867–878. https://doi.org/10.1093/clinids/10.4.867
Huang TD, Bogaerts P, Berhin C, Guisset A, Glupczynski Y (2010) Evaluation of Brilliance ESBL agar, a novel chromogenic medium for detection of extended-spectrum-beta- lactamase-producing Enterobacteriaceae. J Clin Microbiol 48(6):2091–2096. https://doi.org/10.1128/jcm.02342-09
Jacob ME, Keelara S, Aidara-Kane A, Matheu Alvarez JR, Fedorka-Cray PJ (2020) Optimizing a screening protocol for potential extended-spectrum β-lactamase Escherichia coli on MacConkey agar for use in a global surveillance program. J Clin Microbiol. https://doi.org/10.1128/jcm.01039-19
Amjad A, Mirza I, Abbasi S, Farwa U, Malik N, Zia F (2011) Modified hodge test: a simple and effective test for detection of carbapenemase production. Iran J Microbiol 3(4):189–193
Carrër A, Fortineau N, Nordmann P (2010) Use of ChromID extended-spectrum beta-lactamase medium for detecting carbapenemase-producing Enterobacteriaceae. J Clin Microbiol 48(5):1913–1914. https://doi.org/10.1128/jcm.02277-09
Giske CG, Gezelius L, Samuelsen Ø, Warner M, Sundsfjord A, Woodford N (2011) A sensitive and specific phenotypic assay for detection of metallo-β-lactamases and KPC in Klebsiella pneumoniae with the use of meropenem disks supplemented with aminophenylboronic acid, dipicolinic acid and cloxacillin. Clin Microbiol Infect 17(4):552–556. https://doi.org/10.1111/j.1469-0691.2010.03294.x
Yong D, Lee K, Yum JH, Shin HB, Rossolini GM, Chong Y (2002) Imipenem-EDTA disk method for differentiation of metallo-beta-lactamase-producing clinical isolates of Pseudomonas spp and Acinetobacter spp. J Clin Microbiol 40(10):3798–801. https://doi.org/10.1128/jcm.40.10.3798-3801.2002
Tan TY, Ng LS, He J, Koh TH, Hsu LY (2009) Evaluation of screening methods to detect plasmid-mediated AmpC in Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis. Antimicrob Agents Chemother 53(1):146–149. https://doi.org/10.1128/aac.00862-08
Maraskolhe DL, Deotale VS, Mendiratta DK, Narang P (2014) Comparision of Three Laboratory Tests for Detection of AmpC β Lactamases in Klebsiella Species and E Coli. J Clin Diagn Res 8(6):Dc05–08. https://doi.org/10.7860/jcdr/2014/8256.4432
Dashti A, Jadaon M, Abdulsamad A, Dashti H (2009) Heat Treatment of bacteria: a simple method of DNA extraction for molecular techniques. Kuwait Med J 41:117–122
RStudio Team. (2022) RStudio: Integrated Development Environment for R. Boston: RStudio, PBC
Du J, Li P, Liu H, Lü D, Liang H, Dou Y (2014) Phenotypic and Molecular characterization of multidrug resistant Klebsiella pneumoniae isolated from a University Teaching Hospital, China. PloS one 9(4):e95181. https://doi.org/10.1371/journal.pone.0095181
See I, Lessa FC, ElAta OA, Hafez S, Samy K, El-Kholy A et al (2013) Incidence and pathogen distribution of healthcare-associated infections in pilot hospitals in Egypt. Infect Control Hosp Epidemiol 34(12):1281–1288. https://doi.org/10.1086/673985
Ahmed SH, Daef EA, Badary MS, Mahmoud MA, Abd-Elsayed AA (2009) Nosocomial blood stream infection in intensive care units at Assiut University Hospitals (Upper Egypt) with special reference to extended spectrum beta-lactamase producing organisms. BMC Res Notes 2:76. https://doi.org/10.1186/1756-0500-2-76
Abdel-Wahab F, Ghoneim M, Khashaba M, El-Gilany AH, Abdel-Hady D (2013) Nosocomial infection surveillance in an Egyptian neonatal intensive care unit. J Hosp Infect 83(3):196–199. https://doi.org/10.1016/j.jhin.2012.10.017
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG et al (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18(3):268–281. https://doi.org/10.1111/j.1469-0691.2011.03570.x
Adekunle OO (2012) Mechanisms of antimicrobial resistance in bacteria, general approach. Int J Pharma Med Biol Sci 1(2):166–187
Hussain HI, Aqib AI, Seleem MN, Shabbir MA, Hao H, Iqbal Z et al (2021) Genetic basis of molecular mechanisms in β-lactam resistant gram-negative bacteria. Microb Pathog 158:105040. https://doi.org/10.1016/j.micpath.2021.105040
Alhazmi W, Al-Jabri A, Al-Zahrani I (2022) The Molecular characterization of nosocomial carbapenem-resistant Klebsiella pneumoniae co-harboring blaNDM and blaOXA-48 in Jeddah. Microbiol Res 13(4):753–764
Ahmed El-Domany R, El-Banna T, Sonbol F, Abu-Sayedahmed SH (2021) Coexistence of NDM-1 and OXA-48 genes in carbapenem resistant Klebsiella pneumoniae clinical isolates in Kafrelsheikh Egypt. Afr Health Sci 21(2):489–496. https://doi.org/10.4314/ahs.v21i2.2
Khalifa HO, Soliman AM, Ahmed AM, Shimamoto T, Hara T, Ikeda M et al (2017) High carbapenem resistance in clinical gram-negative pathogens isolated in Egypt. Microb Drug Resist 23(7):838–844. https://doi.org/10.1089/mdr.2015.0339
ElMahallawy HA, Zafer MM, Amin MA, Ragab MM, Al-Agamy MH (2018) Spread of carbapenem resistant Enterobacteriaceae at tertiary care cancer hospital in Egypt. Infect Dis (Lond) 50(7):560–4. https://doi.org/10.1080/23744235.2018.1428824
Ragheb SM, Tawfick MM, El-Kholy AA, Abdulall AK (2020) Phenotypic and genotypic features of Klebsiella pneumoniae harboring carbapenemases in Egypt: OXA-48-like carbapenemases as an investigated model. Antibiotics (Basel) 9(12):852. https://doi.org/10.3390/antibiotics9120852
Rand KH, Turner B, Seifert H, Hansen C, Johnson JA, Zimmer A (2011) Clinical laboratory detection of AmpC β-lactamase: does it affect patient outcome? Am J Clin Pathol 135(4):572–576. https://doi.org/10.1309/ajcp7vd0nmamqcwa
Abdel Aal AM, Khalil NO, Rashed H-AG, Abd Elrahman MZ, ElMelegy TH (2021) Genetic detection of AmpC beta-lactamase among gram-negative isolates "A Single Center Experience’’. Egypt J Immunol 28(4):195–205
Sim J, Wright CC (2005) The kappa statistic in reliability studies: use, interpretation, and sample size requirements. Phys Ther 85(3):257–268
Funding
No funding was received for conducting this study.
Author information
Authors and Affiliations
Contributions
AG Conceptualization, Methodology, Validation, Formal analysis, Investigation, Supervision, writing original draft, Review & Editing. GHA Methodology, Validation, Formal analysis, Investigation RE Formal & statistical analysis, Writing, Reviewing and Editing HO Conceptualization, Methodology, Validation, Writing, Review & Editing, Supervision HMA-S Conceptualization, Methodology, Formal analysis, Validation, Writing—Review & Editing, Supervision.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical Approval
The authors confirm that the ethical policies of the journal, as noted on the journal’s author guidelines page, have been adhered to and the appropriate ethical review committee approval has been received. The ethics committee of the Medical Research Institute was granted the following approval numbers for the current study: IORG0008812 and IRB00010526.
Consent to Participate
We obtained informed consent to participate from all participants before starting the study.
Consent for Publication
All study participants gave consent for publication as long as their data were presented anonymously.
We obtained the participants consent before seeking publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gaballah, A., Ali, G.H., Emad, R. et al. Beta-lactam Resistance Profile among Klebsiella pneumoniae Clinical Isolates from Alexandria, Egypt. Curr Microbiol 80, 356 (2023). https://doi.org/10.1007/s00284-023-03479-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03479-7