Abstract
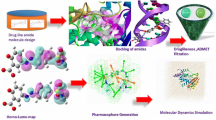
Tuberculosis is the disease which is caused due to the contagion of Mycobacterium tuberculosis. The multidrug resistance Mycobacterium tuberculosis is the main hassle in the treatment of this worldwide health threats. Pantothenate synthase is a legitimate goal for rational drug designing against Mycobacterium tuberculosis. The enzyme is most active in the presence of magnesium or manganese. Marine algal cell wall is rich in sulfated polysaccharides such as fucoidans (brown algae), κ-carrageenans (red algae), and ulvan (green algae) with various favorable biological activities such as anticoagulant, antiviral, antioxidative, anticancer, and immunomodulating activities. In this study, we have modeled binding modes of selected known anti-tubercular compounds and different solvent extract against pantothenate synthase using advanced docking program AutoDock 4.2 tool. In our current study, in silico experiments were carried out to determine if fucoidan, κ-carrageenan, and ulvan sulfated polysaccharides could be a potential target against PANc (pantothenate synthetase), with the goal of identifying potential inhibitors as anti-TB leads targeting PANc for further wet lab validation. Two bioactive compounds were docked to the Mtb pantothenate synthetase protein binding site, with docking scores ranging from − 5.57 to − 2.73. κ-carrageenan had the best pose and docking score, with a Ligand fit score of − 5.815. Ulvan did not dock with the protein. The molecular dynamics simulations were conducted with substrate and ligand bounded fucoidan and κ-carrageenan for 150 ns and the protein Mtb pantothenate synthetase showed a stable conformation in the simulation, with tight amino acid contributions binding to the ligand molecule. RMSD characterizes the conformation and stability of protein ligand complexes, with higher fluctuations indicating low stability and minimal low-level fluctuations indicating equilibration and stability. The graph for RMSF shows significant peaks due to fluctuations in active site regions and other peaks indicating the adaptation of the ligand molecule to the protein binding pocket. From the molecular dynamics study, it is clear that the compounds are having good binding affinity in the active site. The root mean square deviation, root mean square fluctuations, and radius of gyration are supportive evidences which helped us to conclude that the compounds κ-carrageenan and fucoidan are suitable lead molecules for inhibiting pantothenate synthetase. Based on these evidences, the natural compounds from seaweeds can be tested clinically either alone or in combinations against the protein, which could facilitate the designing or the synthesis of new lead molecules as drugs against the tuberculosis.







Similar content being viewed by others
References
Chandra P, Grigsby SJ, Philips JA (2022) Immune evasion and provocation by Mycobacterium tuberculosis. Nat Rev Microbiol 20(12):750–766
World Health Organization (2023) Tuberculosis. https://www.who.int/news-room/fact-sheets/detail/tuberculosis. Accessed 24 March 2023
Alemu A, Bitew ZW, Seid G, Diriba G, Gashu E, Berhe N, Mariam SH, Gumi B (2022) Tuberculosis in individuals who recovered from COVID-19: a systematic review of case reports. PLoS ONE 17(11):e0277807
Global Tuberculosis Report 2019 (World Health Organization, Geneva (2019) Licence: CC BY-NC-SA
Shah YB, Mistry PS, Dhameliya TM, Ranch KM, Boddu SH, Jacob S, Mahalakshmi B, Renukuntla J (2023) Tuberculosis: current treatment options and future scope. In: Shegokar R, Pathak Y (eds) Tubercular drug delivery systems: advances in treatment of infectious diseases. Springer, Cham, pp 59–77
Lange C, Aaby P, Behr MA, Donald PR, Kaufmann SH, Netea MG, Mandalakas AM (2022) 100 years of Mycobacterium bovis bacille Calmette-Guérin. Lancet Infect Dis 22(1):2–12. https://doi.org/10.1016/S1473-3099(21)00403-5
Hawn TR, Day TA, Scriba TJ, Hatherill M, Hanekom WA, Evans TG, Churchyard GJ, Kublin JG, Bekker LG, Self SG (2014) Tuberculosis vaccines and prevention of infection. Microbiol Mol Biol Rev 78(4):650–671
Johnston JC, Cooper R, Menzies D (2022) Chapter 5: Treatment of tuberculosis disease. Can J Respir Crit Care Sleep Med 6(1):66–76
Syed RR, Catanzaro DG, Colman RE, Cooney CG, Linger Y, Kukhtin AV, Holmberg RC, Norville R, Crudu V, Ciobanu N, Codreanu A (2023) Clinical evaluation of the XDR-LFC assay for the molecular detection of isoniazid, rifampin, fluoroquinolone, kanamycin, capreomycin, and amikacin drug resistance in a prospective cohort. J Clin Microbiol 61(3):e01478-e1522
Deshkar AT, Shirure PA, Deshkar A, Shirure P (2022) Bedaquiline: A novel diarylquinoline for multidrug-resistant pulmonary tuberculosis. Cureus. https://doi.org/10.7759/cureus.28519
Dover LG, Coxon GD (2011) Current status and research strategies in tuberculosis drug development: miniperspective. J Med Chem 54(18):6157–6165
Perveen S, Sharma R (2022) Screening approaches and therapeutic targets: the two driving wheels of tuberculosis drug discovery. Biochem Pharmacol 197:114906
Pal A, Kamthania MC, Kumar A (2014) Bioactive compounds and properties of seaweeds—a review. Open Access Library J 1(4):1–7
Pereira L (2018) Therapeutic and nutritional uses of algae. CRC Press, Boca Raton
Knutsen SH, Myslabodski DE, Larsen B, Usov AI (1994) A modified system of nomenclature for red algal galactans. Bot Mar 37:163–169
Conchie J, Percival EG (1950) 167. Fucoidin. Part II. The hydrolysis of a methylated fucoidin prepared from Fucus vesiculosus. J Chem Soc (Resumed) 827–832
Zheng R, Blanchard JS (2001) Steady-state and pre-steady-state kinetic analysis of Mycobacterium tuberculosis pantothenate synthetase. Biochemistry 40(43):12904–12912
Norgan AP, Coffman PK, Kocher JP, Katzmann DJ, Sosa CP (2011) Multilevel parallelization of AutoDock 4.2. J Cheminformatics 3:1–9
Lemkul JA (2018) From proteins to perturbed hamiltonians: a suite of tutorials for the GROMACS-2018 molecular simulation package [article v1. 0]. Living J Comput Mol Sci 1(5068.10):33011
Suresh A, Srinivasarao S, Khetmalis YM, Nizalapur S, Sankaranarayanan M, Sekhar KV (2020) Inhibitors of pantothenate synthetase of Mycobacterium tuberculosis—a medicinal chemist perspective. RSC Adv 10(61):37098–37115
Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, Hutchison GR (2012) Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminformatics 4(1):1–7
Cheeseright T, Mackey M, Rose S, Vinter A (2006) Molecular field extrema as descriptors of biological activity: definition and validation. J Chem Inf Model 46(2):665–676
Muhammed MT, Aki-Yalcin E (2022) Molecular docking: principles, advances, and its applications in drug discovery. Lett Drug Des Discov 19
Stroganov OV, Novikov FN, Stroylov VS, Kulkov V, Chilov GG (2008) Lead finder: an approach to improve accuracy of protein−ligand docking, binding energy estimation, and virtual screening. J Chem Inf Model 48(12):2371–2385
Naz S, Farooq U, Khan S, Sarwar R, Mabkhot YN, Saeed M, Alsayari A, Muhsinah AB, Ul-Haq Z (2021) Pharmacophore model-based virtual screening, docking, biological evaluation and molecular dynamics simulations for inhibitors discovery against α-tryptophan synthase from Mycobacterium tuberculosis. J Biomol Struct Dyn 39(2):610–620
Berendsen HJ, Hess B, Lindahl E, Van Der Spoel D, Mark AE, Groenhof G (2005) GROMACS: fast, flexible, and free. J Comput Chem 26(16):1701–1718
Lindahl E, Hess B, Van Der Spoel D (2001) GROMACS 3.0: a package for molecular simulation and trajectory analysis. Mol Model Annu 7:306–317
Elofsson A, Hess B, Lindahl E, Onufriev A, Van der Spoel D, Wallqvist A (2019) Ten simple rules on how to create open access and reproducible molecular simulations of biological systems. PLoS Comput Biol 15(1):e1006649
Website. PDB available online: http://www.rcsb.org/pdb/
Acknowledgements
The authors wish to thank the researchers supporting Project number RSP-2023R110 at King Saud University Riyadh Saudi Arabia. The authors are grateful to the Science and Engineering Research Board (SERB-Early Career Research Award-ECR/2015/000460), Government of India, New Delhi, for the financial support, and also to the Management of Sathyabama Institute of Science and Technology Chennai, Tamilnadu, for providing the necessary infrastructure facilities to carry out the research work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Arokia Rajan, M., Thirunavukkarasu, R., Joseph, J. et al. Identification of the Seaweed Metabolites as Potential Anti-tubercular Agents Against Human Pantothenate synthetase: An In Silico Approach. Curr Microbiol 80, 318 (2023). https://doi.org/10.1007/s00284-023-03422-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03422-w