Abstract
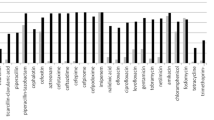
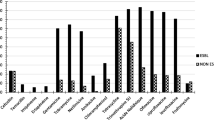
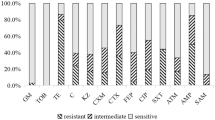
The present study presents phenotypic and molecular characterization of a multidrug-resistant strain of Escherichia coli (Lemef26), belonging to sequence type ST9499 carrying a blaNDM-1 carbapenem resistance gene. The bacterium was isolated from a specimen of Musca domestica, collected in proximity to a hospital in Rio de Janeiro City, Brazil. The strain was identified as E. coli by matrix-assisted laser desorption-ionization time of flight mass spectrometry (Maldi-TOF–MS) and via genotypic analysis (Whole-Genome Sequencing—WGS), followed by phylogenetic analysis, antibiotic resistance profiling (using phenotypic and genotypic methods) and virulence genotyping. Interestingly, the blaNDM-1 was the only resistance determinant detected using a panel of common resistance genes, as evaluated by PCR. In contrast, WGS detected genes conferring resistance to aminoglycosides, fluoroquinolones, quinolones, trimethoprim, beta-lactams, chloramphenicol, macrolides, sulfonamide, tetracycline, lincosamide and streptogramin B. Conjugation experiments demonstrated the transfer of carbapenem resistance, via acquisition of the blaNDM-1 sequence, to a sensitive receptor strain of E. coli, indicating that blaNDM-1 is located on a conjugative plasmid (most likely of the IncA/C incompatibility group, in association with the transposon Tn3000). Phylogenetic analyses placed Lemef26 within a clade of strains exhibiting allelic and environment diversity, with the greatest level of relatedness recorded with a strain isolated from a human source suggesting a possible anthropogenic origin. Analysis of the virulome revealed the presence of fimbrial and pilus genes, including a CFA/I fimbriae (cfaABCDE), common pilus (ecpABCDER), laminin-bind fimbrae (elfADG), hemorrhagic pilus (hcpABC) and fimbrial adherence determinants (stjC) indicates the ability of strain Lemef26 to colonize animal hosts. To the best of our knowledge, this study represents the first report of blaNDM-1 carbapenemase gene in an E. coli strain isolated from M. domestica. In concordance with the findings of previous studies on the carriage of MDR bacteria by flies, the data presented herein provide support to the idea that flies may represent a convenient means (as sentinel animals) for the monitoring of environmental contamination with multidrug-resistant bacteria.


Similar content being viewed by others
References
Taati MM, Mirzaei M, Fazel TMM, Babakhani S, Yeganeh O, Asgharzadeh S et al (2021) The challenge of global emergence of novel colistin-resistant Escherichia coli ST131. Microb Drug Resist 27(11):1513–1524. https://doi.org/10.1089/mdr.2020.0505
Berthe T, Ratajczak M, Clermont O, Denamur E, Petit F (2013) Evidence for coexistence of distinct Escherichia coli populations in various aquatic environments and their survival in estuary water. Appl Environ Microbiol 79(15):4684–4693. https://doi.org/10.1128/AEM.00698-13
Paitan Y (2018) Current trends in antimicrobial resistance of Escherichia coli. Curr Top Microbiol Immunol 416:181–211. https://doi.org/10.1007/82_2018_110
McDanel J, Schweizer M, Crabb V, Nelson R, Samore M, Khader K, Blevins AE, Diekema D, Chiang HY, Nair R, Perencevich E (2017) Incidence of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli and Klebsiella infections in the United States: a systematic literature review. Infect Control Hosp Epidemiol 38(10):1209–1215. https://doi.org/10.1017/ice.2017.156
Luo Y, Luo R, Ding H, Ren X, Luo H, Zhang Y, Ye L, Cui S (2018) Characterization of carbapenem-resistant Escherichia coli isolates through the whole-genome sequencing analysis. Microb Drug Resist 24(2):175–180. https://doi.org/10.1089/mdr.2017.0079
Stapleton AE, Wagenlehner FME, Mulgirigama A, Twynholm M (2020) Escherichia coli resistance to fluoroquinolones in community-acquired uncomplicated urinary tract infection in women: a systematic review. Antimicrob Agents Chemother 64(10):e00862-e920. https://doi.org/10.1128/AAC.00862-20
Ojdana D, Sieńko A, Sacha P, Majewski P, Wieczorek P, Wieczorek A, Tryniszewska E (2018) Genetic basis of enzymatic resistance of E. coli to aminoglycosides. Adv Med Sci 63(1):9–13. https://doi.org/10.1016/j.advms.2017.05.004
Wesolek JL, Wu JY, Smalley CM, Wang L, Campbell MJ (2022) Risk factors for trimethoprim and sulfamethoxazole-resistant Escherichia coli in ED patients with urinary tract infections. Am J Emerg Med 56:178–182. https://doi.org/10.1016/j.ajem.2022.03.052
Ling Z, Yin W, Shen Z, Wang Y, Shen J, Walsh TR (2020) Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J Antimicrob Chemother 75(11):3087–3095. https://doi.org/10.1093/jac/dkaa205
Araújo S, Tacão M, Baraúna R, Ramos R, Silva A, Henriques I (2021) Genome analysis of two multidrug-resistant Escherichia coli O8:H9-ST48 strains isolated from lettuce. Gene 785:145603. https://doi.org/10.1016/j.gene.2021.145603
Hesp A, Ter Braak C, van der Goot J, Veldman K, van Schaik G, Mevius D (2021) Antimicrobial resistance clusters in commensal Escherichia coli from livestock. Zoonoses Public Health 68(3):194–202. https://doi.org/10.1111/zph.12805
Loayza F, Graham JP, Trueba G (2020) Factors obscuring the role of E. coli from domestic animals in the global antimicrobial resistance crisis: an evidence-based review. Int J Environ Res Public Health 17(9):3061. https://doi.org/10.3390/ijerph17093061
Lagerstrom KM, Hadly EA (1948) The under-investigated wild side of Escherichia coli: genetic diversity, pathogenicity and antimicrobial resistance in wild animals. Proc Biol Sci 2021(288):20210399. https://doi.org/10.1098/rspb.2021.0399
Heiden SE, Kurz MSE, Bohnert J, Bayingana C, Ndoli JM, Sendegeya A, Gahutu JB, Eger E, Mockenhaupt FP, Schaufler K (2020) Flies from a tertiary hospital in Rwanda carry multidrug-resistant Gram-negative pathogens including extended-spectrum beta-lactamase-producing E. coli sequence type 131. Antimicrob Resist Infect Control 9(1):34. https://doi.org/10.1186/s13756-020-0696-y
Carramaschi IN, Lopes JCO, Leite JA, Carneiro MT, Barbosa RR, Boas MHV, Rangel K, Chagas TPG, Queiroz MM, Zahner V (2021) Surveillance of antimicrobial resistant bacteria in flies (Diptera) in Rio de Janeiro city. Acta Trop 220:105962. https://doi.org/10.1016/j.actatropica.2021.105962
Transforming our world: the 2030 agenda for sustainable development a/res/70/1. 2015.
de Carvalho CJB, de Mello-Patiu CA (2008) Key to the adults of the most common forensic species of Diptera in South America. Rev Bras entomol 52(3):390–406. https://doi.org/10.1590/S0085-56262008000300012
Patel JB (2017) Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing
van der Zwaluw K, de Haan A, Pluister GN, Bootsma HJ, de Neeling AJ, Schouls LM (2015) The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS ONE 10(3):e0123690. https://doi.org/10.1371/journal.pone.0123690
Sfeir MM, Hayden JA, Fauntleroy KA, Mazur C, Johnson JK, Simner PJ, Das S, Satlin MJ, Jenkins SG, Westblade LF (2019) EDTA-modified carbapenem inactivation method: a phenotypic method for detecting metallo-β-lactamase-producing Enterobacteriaceae. J Clin Microbiol 57(5):e01757-e1818. https://doi.org/10.1128/JCM.01757-18
Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ (2005) Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63(3):219–228. https://doi.org/10.1016/j.mimet.2005.03.018
Kado CI, Liu ST (1981) Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol 145(3):1365–1373. https://doi.org/10.1128/jb.145.3.1365-1373.1981
Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A (2020) Using SPAdes De Novo Assembler. Curr Protoc Bioinform 70(1):102. https://doi.org/10.1002/cpbi.102
Li W, O’Neill KR, Haft DH, DiCuccio M, Chetvernin V, Badretdin A, Coulouris G, Chitsaz F, Derbyshire MK, Durkin AS, Gonzales NR, Gwadz M, Lanczycki CJ, Song JS, Thanki N, Wang J, Yamashita RA, Yang M, Zheng C, Marchler-Bauer A, Thibaud-Nissen F (2021) RefSeq: expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res 49(D1):D1020–D1028. https://doi.org/10.1093/nar/gkaa1105
Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O (2012) Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50(4):1355–1361. https://doi.org/10.1128/JCM.06094-11
Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, Philippon A, Allesoe RL, Rebelo AR, Florensa AF, Fagelhauer L, Chakraborty T, Neumann B, Werner G, Bender JK, Stingl K, Nguyen M, Coppens J, Xavier BB, Malhotra-Kumar S, Westh H, Pinholt M, Anjum MF, Duggett NA, Kempf I, Nykäsenoja S, Olkkola S, Wieczorek K, Amaro A, Clemente L, Mossong J, Losch S, Ragimbeau C, Lund O, Aarestrup FM (2020) ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 75(12):3491–3500. https://doi.org/10.1093/jac/dkaa345
Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H (2014) In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58(7):3895–3903. https://doi.org/10.1128/AAC.02412-14
Liu B, Zheng D, Jin Q, Chen L, Yang J (2019) VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 47(D1):D687–D692. https://doi.org/10.1093/nar/gky1080
Emms DM, Kelly S (2018) STAG: species tree inference from all genes bioRxiv. https://doi.org/10.1101/267914.
Emms DM, Kelly S (2017) STRIDE: species tree root inference from gene duplication events. Mol Biol Evol 34(12):3267–3278. https://doi.org/10.1093/molbev/msx259
Emms DM, Kelly S (2019) OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20(1):238. https://doi.org/10.1186/s13059-019-1832-y
Jolley KA, Bray JE, Maiden MCJ (2018) Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. https://doi.org/10.12688/wellcomeopenres.14826.1
Fuga B, Sellera FP, Cerdeira L, Esposito F, Cardoso B, Fontana H, Moura Q, Cardenas-Arias A, Sano E, Ribas RM, Carvalho AC, Tognim MCB, de Morais MMC, Quaresma AJPG, Santana ÂP, Reis JN, Pilonetto M, Vespero EC, Bonelli RR, Cerqueira AMF, Sincero TCM, Lincopan N (2022) WHO critical priority Escherichia coli as one health challenge for a post-pandemic scenario: genomic surveillance and analysis of current trends in Brazil. Microbiol Spectr 10(2):e0125621. https://doi.org/10.1128/spectrum.01256-21
Meier-Kolthoff JP, Göker M (2019) TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. https://doi.org/10.1038/s41467-019-10210-3
Denamur E, Clermont O, Bonacorsi S, Gordon D (2021) The population genetics of pathogenic Escherichia coli. Nat Rev Microbiol 19(1):37–54. https://doi.org/10.1038/s41579-020-0416-x
Dadashi M, Yaslianifard S, Hajikhani B, Kabir K, Owlia P, Goudarzi M, Hakemivala M, Darban-Sarokhalil D (2019) Frequency distribution, genotypes and prevalent sequence types of New Delhi metallo-β-lactamase-producing Escherichia coli among clinical isolates around the world: a review. J Glob Antimicrob Resist 19:284–293. https://doi.org/10.1016/j.jgar.2019.06.008
Khan AU, Maryam L, Zarrilli R (2017) Structure, genetics and worldwide spread of New Delhi metallo-β-lactamase (NDM): a threat to public health. BMC Microbiol 17(1):101. https://doi.org/10.1186/s12866-017-1012-8
De Oliveira DMP, Forde BM, Kidd TJ, Harris PNA, Schembri MA, Beatson SA, Paterson DL, Walker MJ (2020) Antimicrobial resistance in ESKAPE pathogens. Clin Microbiol Rev 33(3):e00181-e219. https://doi.org/10.1128/CMR.00181-19
Carramaschi IN, Dos S B Ferreira V, Chagas TPG, Corrêa LL, Picão RC, de C Queiroz MM, Rangel K, Jardim R, da Mota FF, Zahner V (2021) Multidrug-resistant Klebsiella quasipneumoniae subsp. similipneumoniae carrying blaNDM-blaCTX-M15 isolated from flies in Rio de Janeiro, Brazil. J Glob Antimicrob Resist 24:1–5. https://doi.org/10.1016/j.jgar.2020.11.021.
Campos JC, da Silva MJ, dos Santos PR, Barros EM, Pereira Mde O, Seco BM, Magagnin CM, Leiroz LK, de Oliveira TG, de Faria-Júnior C, Cerdeira LT, Barth AL, Sampaio SC, Zavascki AP, Poirel L, Sampaio JL (2015) Characterization of Tn3000, a transposon responsible for blaNDM-1 dissemination among enterobacteriaceae in Brazil, Nepal, Morocco, and India. Antimicrob Agents Chemother 59(12):7387–7395. https://doi.org/10.1128/AAC.01458-15
Riley LW (2020) Distinguishing pathovars from nonpathovars: Escherichia coli. Microbiol Spectr. https://doi.org/10.1128/microbiolspec.AME-0014-2020
Sun D, Jin S, Wang J, Wang Z, Fan J, Xu Z, Xu Y, Chen X, Jiao X (2020) Multidrug-resistant Escherichia coli strain isolated from swine in China Harbors mcr-3.1 on a plasmid of the IncX1 type that cotransfers with mcr-1.1. Foodborne Pathog Dis 17(10):597–601. https://doi.org/10.1089/fpd.2019.2769
Watson E, Jeckel S, Snow L, Stubbs R, Teale C, Wearing H, Horton R, Toszeghy M, Tearne O, Ellis-Iversen J, Coldham N (2012) Epidemiology of extended spectrum beta-lactamase E. coli (CTX-M-15) on a commercial dairy farm. Vet Microbiol 154(3–4):339–46. https://doi.org/10.1016/j.vetmic.2011.07.020
Sobur A, Haque ZF, Sabuj AA, Ievy S, Rahman AT, El Zowalaty ME, Rahman T (2019) Molecular detection of multidrug and colistin-resistant Escherichia coli isolated from house flies in various environmental settings. Future Microbiol 14:847–858. https://doi.org/10.2217/fmb-2019-0053
Hickman RA, Agarwal V, Sjöström K, Emanuelson U, Fall N, Sternberg-Lewerin S, Järhult JD (2022) Dissemination of resistant Escherichia coli among wild birds, rodents, flies, and calves on dairy farms. Front Microbiol 13:838339. https://doi.org/10.3389/fmicb.2022.838339
Wetzker W, Pfeifer Y, Wolke S, Haselbeck A, Leistner R, Kola A, Gastmeier P, Salm F (2019) Extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolated from flies in the urban center of Berlin, Germany. Int J Environ Res Public Health 16(9):1530. https://doi.org/10.3390/ijerph16091530
Acknowledgements
Isabel N Carramaschi acknowledges to Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Capes). The authors acknowledge Faperj for financial support (proc n E-26/210.228/2018). The authors acknowledge Dr Alberto R. D´Avila (IOC/FIOCRUZ) for the opportunity to use Illumina MiSeq for Whole-Genome Sequencing.
Funding
Faperj 26/210.228/2018.
Author information
Authors and Affiliations
Contributions
INC: sample collection and perform the experiments, data analysis, manuscript writing. MMdCQ: design the experiments concerning flies capture. FFdM: perform the experiments and data analysis, manuscript writing. VZ: conceive and design the experiments, data analysis, manuscript writing.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to disclose.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Carramaschi, I.N., de C Queiroz, M.M., da Mota, F.F. et al. First Identification of bla NDM-1 Producing Escherichia coli ST 9499 Isolated from Musca domestica in the Urban Center of Rio de Janeiro, Brazil. Curr Microbiol 80, 278 (2023). https://doi.org/10.1007/s00284-023-03393-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03393-y