Abstract
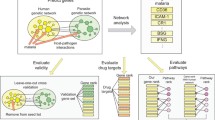
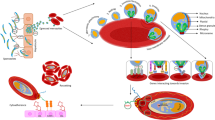
The erythrocyte invasion phase plays a critical role in multiplication, sexual determination, and drug resistance in Plasmodium falciparum. In order to identify the critical genes and pathways in the erythrocyte invasion phase, the gene set (GSE129949) and the RNA-Seq count data for the W2mef strain were used for further analysis. An integrative bioinformatics study was performed to scrutinize genes as potential drug targets. 487 differentially expressed genes (DEGs) with an adjusted P value < 0.001 enriched 47 Gene Ontology (GO) terms that were over-represented based on hyper-geometric analysis P value < 0.01. Protein–Protein interaction network analysis was done using DEGs with higher confidence interactions (PPI score threshold = 0.7). MCODE and cytoHubba apps were utilized to define the hub proteins and rank them based on multiple topological analyses and MCODE scores. Furthermore, Gene Set Enrichment Analysis (GSEA) was carried out by using 322 gene sets from the MPMP database. The genes involved in multiple significant gene sets were determined by leading-edge analysis. Our study identified six genes encoding proteins that could be potential drug targets involved in the erythrocyte invasion phase related to merozoites motility, cell-cycle regulation, G-dependent protein kinase phosphorylation in schizonts, control of microtubule assembly, and sexual commitment. The druggability of those proteins was calculated based on the DCI (Drug Confidence Index) and predicted binding pockets’ values. The protein that showed the best binding pocket value was subjected to deep learning-based virtual screening. The study identified the best small molecule inhibitors in terms of drug-binding score against the proteins for inhibitor identification.





Similar content being viewed by others
References
Cibulskis RE, Alonso P, Aponte J et al (2016) Malaria: global progress 2000–2015 and future challenges. Infect Dis Poverty 5:61. https://doi.org/10.1186/s40249-016-0151-8
Nkumama IN, O’meara WP, Osier FH (2017) Changes in malaria epidemiology in Africa and new challenges for elimination. Trends Parasitol 33:128–140. https://doi.org/10.1016/j.pt.2016.11.006
World Health Organization. World malaria report 2022. World Health Organization, 2022.
Acharya P, Garg M, Kumar P, Munjal A, Raja KD (2017) Host–parasite interactions in human malaria: clinical implications of basic research. Front Microbiol 8:889. https://doi.org/10.3389/fmicb.2017.00889
Zhang M, Wang C, Otto TD, Oberstaller J, Liao X, Adapa SR, Udenze K, Bronner IF, Casandra D, Mayho M, Brown J (2018) Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science 4:360. https://doi.org/10.1126/science.aap7847
WHO Malaria Report (2020)
Miller LH, Baruch DI, Marsh K, Doumbo OK (2002) The pathogenic basis of malaria. Nature 415:673. https://doi.org/10.1038/415673a
Cowman AF, Berry D, Baum J (2012) The cellular and molecular basis for malaria parasite invasion of the human red blood cell. J Cell Biol 198(6):961–971. https://doi.org/10.1083/jcb.201206112
Lopaticki S et al (2011) Reticulocyte and erythrocyte binding-like proteins function cooperatively in invasion of human erythrocytes by malaria parasites. Infect Immun 79:1107–1117. https://doi.org/10.1128/IAI.01021-10
Baum J, Maier AG, Good RT, Simpson KM, Cowman AF (2005) Invasion by P. falciparum merozoites suggests a hierarchy of molecular interactions. PLoS Path 1:e37. https://doi.org/10.1371/journal.ppat.0010037
Campino S et al (2018) A forward genetic screen reveals a primary role for Plasmodium falciparum Reticulocyte Binding Protein Homologue 2a and 2b in determining alternative erythrocyte invasion pathways. PLoS Path 14:e1007436. https://doi.org/10.1371/journal.ppat.1007436
Ahouidi AD et al (2016) Malaria vaccine development: focusing field erythrocyte invasion studies on phenotypic diversity: the West African Merozoite Invasion Network (WAMIN). Trends Parasitol 32:274–283. https://doi.org/10.1016/j.pt.2015.11.009
Persson KE et al (2008) Variation in use of erythrocyte invasion pathways by Plasmodium falciparum mediates evasion of human inhibitory antibodies. J Clin Invest 118:342–351. https://doi.org/10.1172/JCI32138
Wright GJ, Rayner JC (2014) Plasmodium falciparum erythrocyte invasion: combining function with immune evasion. PLoS Pathog 10(3):e1003943. https://doi.org/10.1371/journal.ppat.1003943
Josling GA, Williamson KC, Llinás M (2018) Regulation of sexual commitment and gametocytogenesis in malaria parasites. Annu Rev Microbiol 72:501–519. https://doi.org/10.1146/annurev-micro-090817-062712
Dolan SA, Miller LH, Wellems TE (1990) Evidence for a switching mechanism in the invasion of erythrocytes by Plasmodium falciparum. J Clin Investig 86(2):618–624. https://doi.org/10.1172/JCI114753
Stubbs J, Simpson KM, Triglia T, Plouffe D, Tonkin CJ, Duraisingh MT, Maier AG, Winzeler EA, Cowman AF (2005) Molecular mechanism for switching of P. falciparum invasion pathways into human erythrocytes. Science 309(5739):1384–1387. https://doi.org/10.1126/science.1115257
Nyarko PB, Tarr SJ, Aniweh Y, Stewart LB, Conway DJ, Awandare GA (2020) Investigating a Plasmodium falciparum erythrocyte invasion phenotype switch at the whole transcriptome level. Sci Rep 10(1):1–10. https://doi.org/10.1038/s41598-019-56386-y
Love MI, Anders S, Huber W (2016) DESeq2 vignette. Genome Biol. https://doi.org/10.1101/2022.08.29.505720
Zhu A, Ibrahim JG, Love MI (2019) Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35(12):2084–2092. https://doi.org/10.1093/bioinformatics/bty895
Thissen D, Steinberg L, Kuang D (2002) Quick and easy implementation of the Benjamini-Hochberg procedure for controlling the false positive rate in multiple comparisons. J Educ Behav Stat 27(1):77–83. https://doi.org/10.3102/10769986027001077
Falcon S, Gentleman R (2007) Using GO stats to test gene lists for GO term association. Bioinformatics 23(2):257–258. https://doi.org/10.1093/bioinformatics/btl567
Carlson M (2019) Genome wide annotation for Malaria org.Pf.plasmo.db version 3.8.2.
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ (2019) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47(D1):D607–D613. https://doi.org/10.1093/nar/gky1131
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY (2014) cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. https://doi.org/10.1186/1752-0509-8-S4-S11
Bader GD, Hogue CW (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4(1):1–27. https://doi.org/10.1186/1471-2105-4-2
Agüero F, Al-Lazikani B, Aslett M, Berriman M, Buckner FS, Campbell RK, Carmona S, Carruthers IM, Chan AE, Chen F, Crowther GJ (2008) Genomic-scale prioritization of drug targets: the TDR Targets database. Nat Rev Drug Discovery 7(11):900–907. https://doi.org/10.1038/nrd2684
Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. The proteomics protocols handbook, pp 571–607. https://doi.org/10.1385/1-59259-890-0:571.
Krogh A, Larsson B, Von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305(3):567–580. https://doi.org/10.1006/jmbi.2000.4315
Ginsburg H, Abdel-Haleem AM (2016) Malaria parasite metabolic pathways (MPMP) upgraded with targeted chemical compounds. Trends Parasitol 32(1):7–9. https://doi.org/10.1016/j.pt.2015.10.003
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102(43):15545–15550. https://doi.org/10.1073/pnas.0506580102
Merico D, Isserlin R, Stueker O, Emili A, Bader GD (2010) Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 5(11):e13984. https://doi.org/10.1371/journal.pone.0013984
Volkamer A, Kuhn D, Rippmann F, Rarey M (2012) DoGSiteScorer: a web server for automatic binding site prediction, analysis and druggability assessment. Bioinformatics 28(15):2074–2075. https://doi.org/10.1093/bioinformatics/bts310
Huang K, Fu T, Glass LM, Zitnik M, Xiao C, Sun J (2020) DeepPurpose: a deep learning library for drug–target interaction prediction. Bioinformatics 36(22–23):5545–5547. https://doi.org/10.1093/bioinformatics/btaa1005
Gilson MK et al (2016) BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. https://doi.org/10.1093/nar/gkv1072
Mahmud S, Paul GK, Biswas S, Kazi T, Mahbub S, Mita MA, Afrose S, Islam A, Ahaduzzaman S, Hasan MR, Shimu MSS (2022) phytochemdb: a platform for virtual screening and computer-aided drug designing. Database. https://doi.org/10.1093/database/baac002
QikProp (2009) QikProp, version 3.2.
Van Biljon R, Van Wyk R, Painter HJ, Orchard L, Reader J, Niemand J, Llinás M, Birkholtz LM (2019) Hierarchical transcriptional control regulates Plasmodium falciparum sexual differentiation. BMC Genomics 20(1):1–6. https://doi.org/10.1186/s12864-019-6322-9
Walzer KA, Kubicki DM, Tang X, Chi JT (2018) Single-cell analysis reveals distinct gene expression and heterogeneity in male and female Plasmodium falciparum gametocytes. Msphere 3(2):e00130-e218. https://doi.org/10.1128/mSphere.00130-18
Martins RM, Macpherson CR, Claes A, Scheidig-Benatar C, Sakamoto H, Yam XY, Preiser P, Goel S, Wahlgren M, Sismeiro O, Coppée JY (2017) An ApiAP2 member regulates expression of clonally variant genes of the human malaria parasite Plasmodium falciparum. Sci Rep 7(1):1. https://doi.org/10.1038/s41598-017-12578-y
Ferreira JL, Heincke D, Wichers JS, Liffner B, Wilson DW, Gilberger TW (2020) The dynamic roles of the inner membrane complex in the multiple stages of the malaria parasite. Front Cell Infect Microbiol. https://doi.org/10.3389/fcimb.2020.611801
More KR, Kaur I, Giai Gianetto Q, Invergo BM, Chaze T, Jain R, Huon C, Gutenbrunner P, Weisser H, Matondo M, Choudhary JS (2020) Phosphorylation-Dependent assembly of a 14–3-3 mediated signaling complex during Red blood cell invasion by Plasmodium falciparum merozoites. MBio 11(4):e01287-e1320. https://doi.org/10.1128/mBio.01287-20
Anand G, Reddy KS, Pandey AK, Mian SY, Singh H, Mittal SA, Amlabu E, Bassat Q, Mayor A, Chauhan VS, Gaur D (2016) A novel Plasmodium falciparum rhoptry associated adhesin mediates erythrocyte invasion through the sialic-acid dependent pathway. Sci Rep 6(1):1–7. https://doi.org/10.1038/srep29185
Arumugam TU, Takeo S, Yamasaki T, Thonkukiatkul A, Miura K, Otsuki H et al (2011) Discovery of GAMA, a Plasmodium falciparum merozoite micronemal protein, as a novel blood-stage vaccine candidate antigen. Infect Immun 79(11):4523–4532. https://doi.org/10.1128/IAI.05412-11
Acknowledgements
The authors would like to acknowledge the Bioinformatics lab facility of the School of Biotechnology, KIIT Deemed to be University, Bhubaneswar during the course of work.
Funding
The project work is supported by the SERB-MATRICS project (MTR/2021/000191) from Department of Science and Technology, Govt. of India awarded to Dr. Rajani Kanta Mahapatra.
Author information
Authors and Affiliations
Contributions
MMK has developed the bioinformatics pipeline and performed the RNA-Seq data analysis. MMA contributed towards the druggability and virtual screening results. RKM supervised the whole study and corrected the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that no conflict of interest exists.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kazan, M.M., Asmare, M.M. & Mahapatra, R.K. Identification of Potential Drug Targets in Erythrocyte Invasion Pathway of Plasmodium falciparum. Curr Microbiol 80, 165 (2023). https://doi.org/10.1007/s00284-023-03282-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03282-4