Abstract
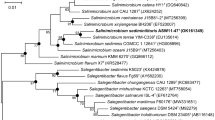
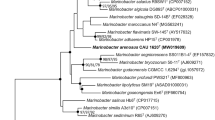
An actinobacterium, designated as SYSU T00001T, was isolated from a tidal flat sediment sample from Guangdong province, China. Cells were Gram-stain-positive, aerobic, motile and short rod-shaped. Colonies on marine agar 2216 were smooth, yellow-pigmented, and circular with low convexity. The isolate was able to grow at the temperature range 4–37 °C (optimum 30 °C), at pH 4.0–10.0 (optimum 7.0) and in the presence of 0–10% (w/v) NaCl. The major menaquinones were MK-11 and MK-10. The cell wall contained alanine, glutamic acid, lysine and ornithine. The major fatty acids were C19:0 cyclo ω8c (35.7%) and anteiso C15:0 (26.0%). The polar lipids consisted of one diphosphatidyl glycerol, one unidentified glycolipid and one unknown lipid. Whole genome sequencing of strain SYSU T00001T revealed 2,837,702 bp with a DNA G + C content of 67.8%. Phylogenetic analyses clearly demonstrated that strain SYSU T00001T belonged to the genus Salinibacterium, and the highest 16S rRNA gene similarity to Salinibacterium hongtaonis 194T (97.8%). The ANI and dDDH values of strain SYSU T00001T relative to Salinibacterium hongtaonis 194T were 74.5% and 19.5%, respectively. According to our data, strain SYSU T00001T represents a novel species of the genus Salinibacterium, for which the name Salinibacterium sedimenticola sp. nov. is proposed, the type strain is SYSU T00001T (= GDMCC 1.3283T = KCTC 49758T).


Similar content being viewed by others
Data Availability
The 16S rRNA gene sequence and draft genome of strain SYSU T00001T have been deposited in GenBank/EMBL/DDBJ.
References
Han SK, Nedashkovskaya OI, Mikhailov VV, Kim SB, Bae KS (2003) Salinibacterium amurskyense gen. nov., sp. nov., a novel genus of the family Microbacteriaceae from the marine environment. Int J Syst Evol Microbiol 53(6):2061–2066. https://doi.org/10.1099/ijs.0.02627-0
Zhang DC, Liu HC, Xin YH, Yu Y, Zhou PJ, Zhou YG (2008) Salinibacterium xinjiangense sp. nov., a psychrophilic bacterium isolated from the China No. 1 glacier. Int J Syst Evol Microbiol 58(12):2739–2742. https://doi.org/10.1099/ijs.0.65802-0
Li J, Lu S, Jin D, Yang J, Lai XH, Zhang G et al (2019) Salinibacterium hongtaonis sp. nov., isolated from faeces of Tibetan antelope (Pantholops hodgsonii) on the Qinghai-Tibet Plateau. Int J Syst Evol Microbiol 69(4):1093–1098. https://doi.org/10.1099/ijsem.0.003277
Gao S (2019) Geomorphology and sedimentology of tidal flats. Coastal Wetlands 10:359–381. https://doi.org/10.1016/b978-0-444-63893-9.00010-1
Wang Y, Sheng HF, He Y, Wu JY, Jiang YX, Tam NF et al (2012) Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl Environ Microbiol 78(23):8264–8271. https://doi.org/10.1128/AEM.01821-12
Li S, Shi L, Lian WH, Lin ZL, Lu CY, Xu L et al (2021) Arenibaculum pallidiluteum gen. nov., sp. nov., a novel bacterium in the family Azospirillaceae, isolated from desert soil, and reclassification of Skermanella xinjiangensis to a new genus Deserticella as Deserticella xinjiangensis comb nov., and transfer of the genera Indioceanicola and Oleisolibacter from the family Rhodospirillaceae to the family Azospirillaceae. Int J Syst Evol Microbiol 71(7). https://doi.org/10.1099/ijsem.0.004874
Li S, Dong L, Lian WH, Lin ZL, Lu CY, Xu L et al (2021) Exploring untapped potential of Streptomyces spp. in Gurbantunggut Desert by use of highly selective culture strategy. Sci Total Environ 790:148235. https://doi.org/10.1016/j.scitotenv.2021.148235
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16(2):111–20. https://doi.org/10.1007/BF01731581
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17(6):368–376. https://doi.org/10.1007/bf01734359
Fitch WM (1971) Toward defining the course of evolution: Minimum change for a specific tree topology. Syst Zoology 20(4):406–416. https://doi.org/10.2307/2412116
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. https://doi.org/10.1093/molbev/msy096
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25(24):4876–4882. https://doi.org/10.1093/nar/25.24.4876
Harrison PG, Strulo B (2000) SPADES-a process algebra for discrete event simulation. J Log Comput 10:3–41
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25(7):1043–1055. https://doi.org/10.1101/gr.186072.114
Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M (2016) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44(D1):D457-462. https://doi.org/10.1093/nar/gkv1070
Galperin MY, Kristensen DM, Makarova KS, Wolf YI, Koonin EV (2019) Microbial genome analysis: the COG approach. Brief Bioinform 20(4):1063–1070. https://doi.org/10.1093/bib/bbx117
Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J (2016) JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32(6):929–931. https://doi.org/10.1093/bioinformatics/btv681
Lu CY, Dong L, Wang D, Li S, Fang BZ, Han MX et al (2022) Dongia deserti sp. nov., Isolated from the Gurbantunggut Desert Soil. Curr Microbiol 79(11):342. https://doi.org/10.1007/s00284-022-03051-9
Bowers RM, Kyrpides NC, Stepanauska R, Harmon-Smith M, Doud D, Reddy TBK et al (2018) Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 35(8):725–731. https://doi.org/10.1038/nbt.3893
Salam N, Jiao JY, Zhang XT, Li WJ (2020) Update on the classification of higher ranks in the phylum Actinobacteria. Int J Syst Evol Microbiol 70(2):1331–1355. https://doi.org/10.1099/ijsem.0.003920
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Wu M, Scott AJ (2012) Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 28(7):1033–1034. https://doi.org/10.1093/bioinformatics/bts079
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17(4):540–552. https://doi.org/10.1093/oxfordjournals.molbev.a026334
Buck JD (1982) Nonstaining (KOH) method for determination of gram reactions of marine bacteria. Appl Environ Microbiol 44(4):992–923. https://doi.org/10.1128/aem.44.4.992-993.1982
Xu P, Li WJ, Tang SK, Zhang YQ, Chen GZ, Chen HH et al (2005) Naxibacter alkalitolerans gen. nov., sp. nov,. a novel member of the family ‘Oxalobacteraceae’ isolated from China. Int J Syst Evol Microbiol 55(3):1149–1153. https://doi.org/10.1099/ijs.0.63407-0
Narsing Rao MP, Dong ZY, Kan Y, Zhang K, Fang BZ, Xiao M et al (2020) Description of Paenibacillus antri sp. nov. and Paenibacillus mesophilus sp. nov., isolated from cave soil. Int J Syst Evol Microbiol 70(2):1048–1054. https://doi.org/10.1099/ijsem.0.003870
Jacobs JM, Pesce C, Lefeuvre P, Koebnik R (2015) Comparative genomics of a cannabis pathogen reveals insight into the evolution of pathogenicity in Xanthomonas. Front Plant Sci 6:431
Gonzalez C, Gutierrez C, Ramirez C (1978) Halobacterium vallismortis sp. nov. An amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can J Microbiol 24(6):710–715. https://doi.org/10.1139/m78-119
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100(2):221–230. https://doi.org/10.1099/00221287-100-2-221
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A et al (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Meth 2(5):233–241. https://doi.org/10.1016/0167-7012(84)90018-6
Kroppenstedt RM (1982) Separation of bacterial menaquinones by HPLC using reverse phase (RP18) and a silver loaded ion exchanger as stationary phases. J Liq Chromatogr 5(12):2359–2367. https://doi.org/10.1080/01483918208067640
Tamaoka J (1986) Analysis of bacterial menaquinone mixtures by reverse-phase high-performance liquid chromatography. Methods Enzymol 123:251–256. https://doi.org/10.1016/S0076-6879(86)23028-1
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. USFCC Newsl 20:16
Komagata K, Suzuki K (1987) Lipid and cell wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Minnikin DE, Collins MD, Goodfellow M (1979) Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Bacteriol 47(1):87–95. https://doi.org/10.1111/j.1365-2672.1979.tb01172.x
Collins M, JONES D, (1980) Lipids in the classification and identification of coryneform bacteria containing peptidoglycans based on 2, 4-diaminobutyric acid. J Appl Bacteriol 48(3):459–470. https://doi.org/10.1111/j.1365-2672.1980.tb01036.x
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106(45):19126–19131. https://doi.org/10.1073/pnas.0906412106
Kim M, Oh HS, Park SC, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64(Pt 2):346–351. https://doi.org/10.1099/ijs.0.059774-0
Acknowledgements
The authors are grateful to Professor Yu-Guang Zhou (CGMCC, China) for providing the reference type strain.
Funding
This research was financially supported by National Science and Technology Fundamental Resources Investigation Program of China (Grant No. 2019FY100700), National Natural Science Foundation of China (Nos: 32270076 and 32000005) and the Key-Area Research and Development Program of Guangdong Province (No. 2022B0202110001). WJ Li was also supported by Introduction project of high-level talents in Xinjiang Uygur Autonomous Region.
Author information
Authors and Affiliations
Contributions
BZF and WJL jointly conceived the study. Authors CYL, LD, SL, WHL, ZLL, ZHZ and LG performed sampling, isolation and identification. CYL, LD and SL drafted the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declared that there is no conflict of interest.
Ethical Approval
No animals or human participants were included in the present study.
Consent for Publication
All the authors agree to submit for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lu, CY., Dong, L., Li, S. et al. Salinibacterium sedimenticola sp. nov., Isolated from Tidal Flat Sediment. Curr Microbiol 80, 142 (2023). https://doi.org/10.1007/s00284-023-03243-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03243-x