Abstract
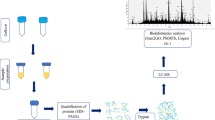
Members of the Enterobacter genus include many pathogenic microbes of humans and plants, secrete proteins that contribute to the interactions of bacteria and their environment. Therefore, understanding of secreted proteins is vital to understand bacterial physiology and behavior. Here, we explored the secretome of an environmental isolate Enterobacter sp. S-16 by nanoLC-MS/MS and identified 572 proteins in the culture supernatant. Gene ontology (GO) analysis indicated that proteins were related to biological processes, molecular as well as cellular functions. The majority of the identified proteins are involved in microbial metabolism, chemotaxis & motility, flagellar hook-associated proteins, biosynthesis of antibiotics, and molecular chaperones to assist the protein folding. Bioinformatics analysis of the secretome revealed the presence of type I and type VI secretion system proteins. Presence of these diverse secretion system proteins in Enterobacter sp. S-16 are likely to be involved in the transport of various proteins including nutrient acquisition, adhesion, colonization, and homeostasis maintenance. Among the secreted bacterial proteins with industrial importance, lignocellulolytic enzymes play a major role, therefore, we analyzed our secretome results for any presence of glycoside hydrolases (GHs) and other hydrolytic enzymes (CAZymes). Overall, the secreted proteins may be considered an attractive reservoir of potential antigens for drug development, diagnostic markers, and other biomedical applications.





Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Code Availability
Not applicable.
References
Ferry A, Plaisant F, Ginevra C et al (2020) Enterobacter cloacae colonization and infection in a neonatal intensive care unit: retrospective investigation of preventive measures implemented after a multiclonal outbreak. BMC Infect Dis 20:682. https://doi.org/10.1186/s12879-020-05406-8
Davin-Regli A, Lavigne JP, Pagès JM (2019) Enterobacter spp.: update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin Microbiol Rev 32:e00002-19. https://doi.org/10.1128/CMR.00002-19
Rice LB (2008) Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis 197:1079–1081. https://doi.org/10.1086/533452
Mezzatesta ML, Gona F, Stefani S (2012) Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol 7(7):887–902. https://doi.org/10.2217/fmb.12.61
Maffei B, Francetic O, Subtil A (2017) Tracking proteins secreted by bacteria: what’s in the toolbox? Front Cell Infect Microbiol 31(7):221. https://doi.org/10.3389/fcimb.2017.00221
Gagic D, Ciric M, Wen WX, Ng F, Rakonjac J (2016) Exploring the secretomes of microbes and microbial communities using filamentous phage display. Front Microbiol 7:927. https://doi.org/10.3389/fmicb.2016.00429
Liang X, Pei T-T, Li H, Zheng H-Y, Luo H, Cui Y et al (2021) VgrG-dependent effectors and chaperones modulate the assembly of the type VI secretion system. PLoS Pathog 17(12):e1010116. https://doi.org/10.1371/journal.ppat.1010116
Costa T, Felisberto-Rodrigues C, Meir A, Prevost Marie S, Redzej A, Trokter M, Waksman G (2015) Secretion systems in gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol 13(6):343–359. https://doi.org/10.1038/nrmicro3456
Schwarz S, Hood RD, Mougous JD (2010) What is type VI secretion doing in all those bugs? Trends Microbiol 18:531–537. https://doi.org/10.1016/j.tim.2010.09.001
Schwarz S, West TE, Boyer F, Chiang W-C, Carl MA et al (2010) Burkholderia Type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog 6(8):e1001068. https://doi.org/10.1371/journal.ppat.1001068
Foster TJ, Geoghegan JA, Ganesh VK, Höök M (2014) Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol 12:49–62. https://doi.org/10.1038/nrmicro3161
Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. https://doi.org/10.1093/nar/gkt1178
López-Mondéjar R, Zühlke D, Větrovský T, Becher D, Ríedel K, Baldrian P (2016) Decoding the complete arsenal for cellulose and hemicellulose deconstruction in the highly efficient cellulose decomposer Paenibacillus O199. Biotechnol Biofuels 9:104. https://doi.org/10.1186/s13068-016-0518-x
Herrera LM, Braña V, Laura Franco Fraguas LF, Castro-Sowinsk S (2019) Characterization of the cellulase-secretome produced by the Antarctic bacterium Flavobacterium sp. AUG42. Microbiol Res. https://doi.org/10.1016/j.micres.2019.03.009
Piccinni FE, Ontañon OM, Ghio S, Sauka DH, Talia PM, Rivarola ML, Valacco MP, Campos E (2018) Secretome profile of Cellulomonas sp. B6 growing on lignocellulosic substrates. J Appl Microbiol 126(3):811–825. https://doi.org/10.1111/jam.14176
Islam MS, Haque MS, Islam MM, Emdad EM, Halim A, Hossen QM, Hossain Z, Ahmad B, Rahim S, Rahman S, Alam M, Hou S, Wan X, Saito JA, Alam M (2012) Tools to kill: genome of one of the most destructive plant pathogenic fungi Macrophomina phaseolina. BMC Genom 13:493. https://doi.org/10.1186/1471-2164-13-493
Zubair M, Khan F, Menghwar H, Faisal M, Ashraf M, Rasheed M et al (2020) Progresses on bacterial secretomes enlighten research on Mycoplasma secretome. Microb Pathog 144:104–160. https://doi.org/10.1016/j.micpath.2020.104160
Bumann D, Aksu S, Wendland M, Janek K, Zimny-Arndt U, Sabarth N, Meyer TF, Jungblut PR (2002) Proteome analysis of secreted proteins of the gastric pathogen Helicobacter pylori. Infect Immun 70:3396–3403. https://doi.org/10.1128/IAI.70.7.3396-3403.2002
Trost SG, Mciver KL, Pate RR (2005) Conducting accelerometer-based activity assessments in field-based research. Med Sci Sport Exer 37:S531–S543. https://doi.org/10.1249/01.mss.0000185657.86065.98
Malen H, Berven FS, Fladmark KE, Wiker HG (2007) Comprehensive analysis of exported proteins from Mycobacterium tuberculosis H37Rv. Proteomics 7:1702–1718. https://doi.org/10.1002/pmic.200600853
Zhao X, Palma Medina LM, Stobernack T, Glasner C, de Jong A, Utari P, Setroikromo R, Quax WJ, Otto A, Becher D, Buist G, van Dijl JM (2019) Exoproteome heterogeneity among closely related Staphylococcus aureus t437 isolates and possible implications for virulence. J Proteome Res 18:2859–2874. https://doi.org/10.1021/acs.jproteome.9b00179
Indrelid SS, Mathiesen G, Jacobsen G, Lea T, Kleiveland CR (2014) Computational and experimental analysis of the secretome of Methylococcus capsulatus (Bath). PLoS One 9:e114476. https://doi.org/10.1371/journal.pone.0114476
Saitz W, Montero DA, Pardo M, Araya D, De la Fuente M, Hermoso MA, Farfán MJ, Ginard D, Rosselló-Móra R, Rasko DA, Del Canto F, Vidal RM (2022) Int J Mol Sci 23(16):9005. https://doi.org/10.3390/ijms23169005
Méndez-Olvera ET, Bustos-Martínez JA, López-Vidal Y, Verdugo-Rodríguez A, Martínez-Gómez D (2016) Jundishapur J Microbiol 9(10):e35591. https://doi.org/10.5812/jjm.35591
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Chemonges S, Gupta R, Mills PC et al (2016) Characterization of the circulating acellular proteome of healthy sheep using LC-MS/MS-based proteomics analysis of serum. Proteome Sci 15:11. https://doi.org/10.1186/s12953-017-0119-z
Nilsson J (2016) Liquid chromatography-tandem mass spectrometry-based fragmentation analysis of glycopeptides. Glycoconj J 33:261–272. https://doi.org/10.1007/s10719-016-9649-3
Gengenbacher M, Mouritsen J, Schubert OT, Aebersold R, Kaufmann SH (2014) Mycobacterium tuberculosis in the proteomics era. Mol Gene Myco. https://doi.org/10.1128/microbiolspec
Liaci AM, Förster F (2021) Take me home, protein roads: structural insights into signal peptide interactions during ER translocation. Int J Mol Sci 22:11871. https://doi.org/10.3390/ijms222111871
Czech L, Mais CN, Kratzat H et al (2022) Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp. Nat Commun 13:1069. https://doi.org/10.1038/s41467-022-28675-0
Gerlach RG, Hensel M (2007) Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens. Int J Med Microbiol 297:401–415. https://doi.org/10.1016/j.ijmm.2007.03.017
Zoued A, Brunet YR, Durand E, Aschtgen MS, Logger L, Douzi B, Journet L, Cambillau C, Cascales E (2014) Architecture and assembly of the type VI secretion system. Biochim Biophys Acta 1843:1664–1673. https://doi.org/10.1016/j.bbamcr.2014.03.018
Lin J, Huang S, Zhang Q (2002) Outer membrane proteins: key players for bacterial adaptation in host niches. Microb Infect 4:325–331. https://doi.org/10.1016/s1286-4579(02)01545-9
Mecsas J, Welch R, Erickson JW, Gross CA (1995) Identification and characterization of an outer membrane protein, OmpX, in Escherichia coli that is homologous to a family of outer membrane proteins including Ail of Yersinia enterocolitica. J Bacteriol 177:799–804. https://doi.org/10.1128/jb.177.3.799-804
Rooke JL, Icke C, Wells TJ, Rossiter AE, Browning DF, Morris FC, Leo JC, Schütz MS, Autenrieth IB, Cunningham AF, Linke D, Henderson IR (2021) BamA and BamD are essential for the secretion of trimeric autotransporter adhesins. Front Microbiol 12:628879. https://doi.org/10.3389/fmicb.2021.628879
Hartl FU (1996) Molecular chaperones in cellular protein folding. Nature 381:571–580. https://doi.org/10.1038/381571a0
Corigliano MG, Sander VA, Sánchez López EF, Ramos Duarte VA, Mendoza Morales LF, Angel SO, Clemente M (2021) Heat shock proteins 90 kDa: Immunomodulators and adjuvants in vaccine design against infectious diseases. Front Bioeng Biotechnol 8:622186. https://doi.org/10.3389/fbioe.2020.622186
Kumari K, Sharma PK, Aggarwal Y, Singh RP (2022) Secretome analysis of an environmental isolate Enterobacter sp. S-33 identifies proteins related to pathogenicity. Arch Microbiol 204(11):662. https://doi.org/10.1007/s00203-022-03277-y
Schramm FD, Kristen Schroeder K, Jonas K (2020) Protein aggregation in bacteria. FEMS Microbiol Rev 44(1):154–172. https://doi.org/10.1093/femsre/fuz026
Haiko J, Westerlund-Wikström B (2013) The role of the bacterial flagellum in adhesion and virulence. MDPI Biol 2:1242–1267. https://doi.org/10.3390/biology2041242
Ebner P, Luqman A, Reichert S, Hauf K, Popella P, Forchhammer K, Otto M, Götz F (2017) Non-classical protein excretion is boosted by PSM alpha-induced cell leakage. Cell Rep 20:1278–1286. https://doi.org/10.1016/j.celrep.2017.07.045
Götz F, Yu W, Dube L, Prax M, Ebner P (2015) Excretion of cytosolic proteins (ECP) in bacteria. Int J Med Microbiol 305(2):230–237. https://doi.org/10.1016/j.ijmm.2014.12.021
Ebner P, Prax M, Nega M, Koch I, Dube L, Yu W, Rinker J, Popella P, Flotenmeyer M, Götz F (2015) Excretion of cytoplasmic proteins (ECP) in Staphylococcus aureus. Mol Microbiol 97:775–789. https://doi.org/10.1111/mmi.13065
Ebner P, Rinker J, Götz F (2016) Excretion of cytoplasmic proteins in Staphylococcus is most likely not due to cell lysis. Curr Genet 62:19–23. https://doi.org/10.1007/s00294-015-0504-z
Dragone G, Kerssemakers AAJ, Driessen JLSP, Yamakawa CK, Brumano LP, Mussatto SI (2020) Innovation and strategic orientations for the development of advanced biorefineries. Bioresour Technol 302:122847. https://doi.org/10.1016/j.biortech.2020.122847
Ren Z, You W, Wu S, Poetsch A, Xu C (2019) Secretomic analyses of Ruminiclostridium papyrosolvens reveal its enzymatic basis for lignocellulose degradation. Biotech Biofuels 12:1–14. https://doi.org/10.1186/s13068-019-1522-8
Wakarchuk WW, Brochu D, Foote S, Robotham A, Saxena H, Erak T, Kelly J (2016) Proteomic analysis of the secretome of Cellulomonas fimi ATCC 484 and Cellulomonas flavigena ATCC 482. PLoS One 11:e0151186. https://doi.org/10.1371/journal.pone.0151186
Onyango SO, Juma J, De Paepe K, Van de Wiele T (2021) Oral and gut microbial carbohydrate-active enzymes landscape in health and disease. Front Microbiol 12:653448. https://doi.org/10.3389/fmicb.2021.653448
Aakko J, Pietilä S, Toivonen R, Rokka A, Mokkala K, Laitinen K et al (2020) A carbohydrate-active enzyme (CAZy) profile links successful metabolic specialization of Prevotella to its abundance in gut microbiota. Sci Rep 10:12411. https://doi.org/10.1038/s41598-020-69241-2
Acknowledgements
The study was supported by a grant from Department of Biotechnology, Government of India.
Funding
The study was funded by Ramalingswami Re-entry Fellowship, Department of Biotechnology, Government of India (Grant Number BT/RLF 2020-21).
Author information
Authors and Affiliations
Contributions
KK and PKS analyzed the secretome data. RPS supervised the work and wrote the original draft.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no competing interests.
Ethical Approval
This article does not contain any studies with human participants or animal performed by any of the authors.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
284_2023_3197_MOESM1_ESM.docx
Supplementary file1 (DOCX 191 kb)—Neighbor-joining phylogenetic tree based on 16S rRNA sequence. Tree shows phylogenetic relationship between Enterobacter sp. S-16 and other type strains of Enterobacter. Supplementary Fig. 1 Neighbor-joining phylogenetic tree based on 16S rRNA sequence. Tree shows phylogenetic relationship between Enterobacter sp. S-16 and other type strains of Enterobacter. The tree was constructed by using MEGA 7. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test with 1000 replicates
284_2023_3197_MOESM2_ESM.xlsx
Supplementary file2 (XLSX 202 kb)—List of identified secretome proteins (572) have been provided in Supplementary File 2
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumari, K., Sharma, P.K. & Singh, R.P. Unraveling the Virulence Factors and Secreted Proteins of an Environmental Isolate Enterobacter sp. S-16. Curr Microbiol 80, 88 (2023). https://doi.org/10.1007/s00284-023-03197-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03197-0