Abstract
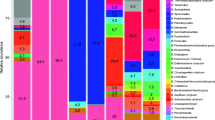
Thermophiles inhabiting high temperatures are considered primitive microorganisms on early Earth. In this regard, several works have demonstrated microbial community composition in geothermal environments. Despite that, studies on hot springs located in the Indian subcontinent viz., Surajkund in the district Hazaribag, Jharkhand; Bakreshwar in the district Birbhum, West Bengal; Tantloi in the district Dumka, and Sidpur in the district Pakur, Jharkhand are scanty. Nonetheless, the metagenomic analysis of these hot springs showed significant differences in the predominant phyla corresponding to geochemical properties. The Chloroflexi, Proteobacteria, Actinobacteria, Deinococcus-Thermus, and Firmicutes were dominant phyla in all the samples. In contrast, Meiothermus was more in comparatively low-temperature hot springs. In addition, archaeal phyla, Euryarchaeota, Candidatus Bathyarchaeota, and Crenarchaeota were predominant in all samples. The canonical correspondence analysis (CCA) showed the abundance of Deinococcus, Thermus, Pyrobaculum, Kocuria, and Geodermatophilus positively correlated with the aqueous concentration of sulfate, fluoride, and argon in relatively high-temperature (≥ 72 °C) hot springs. However, at a lower temperature (≤ 63 °C), Thermodesulfovibrio, Caldilinea, Chloroflexus, Meiothermus, and Tepidimonas are positively correlated with the concentration of zinc, iron, and dissolved oxygen. Further, hierarchical clustering exhibits variations in its functional attributes depending on the temperature gradients. Metagenome analysis predicted carbon, methane, sulfur, and nitrogen metabolism genes, indicating a wide range of bacteria and archaea habitation in these hot springs. In addition, identified several genes encode polyketide biosynthesis pathways. The present study described the microbial community composition and function in the tropical hot springs and their relationship with the environmental variables.





Similar content being viewed by others
Data Availability
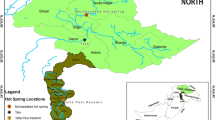
The raw shotgun sequence reads are available in the NCBI-SRA database under the following accession numbers: SRR8368399 (sample-1, Surajkund main source), SRR8369092 (sample-2, Surajkund surrounding area), SRR8369165 (sample-3, Bakreshwar), SRR8369793 (sample-4, Tantloi main source), SRR8369840 (sample-5, Tantloi surrounding area), and SRR8371833 (sample-6, Sidpur).
References
Renaut RW, Jones B (2011) Hydrothermal environments, terrestrial. In: Reiner J, Thiel V (eds) Encyclopedia of Geobiology. Springer, Amsterdam, pp 467–479. https://doi.org/10.1007/978-1-4020-9212-1_114
Power JF, Carere CR, Lee CK, Wakerley GLJ, Evans DW et al (2018) Microbial biogeography of 925 geothermal springs in New Zealand. Nat Commun 9:2876. https://doi.org/10.1038/s41467-018-05020-y
Sriaporn C, Campbell KA, Millan M, Ruff SW, Van Kranendonk MJ et al (2020) Stromatolitic digitate sinters form under wide-ranging physicochemical conditions with diverse hot spring microbial communities. Geobiology 18:619–640. https://doi.org/10.1111/gbi.12395
Meyer-Dombard DR, Amend JP (2014) Geochemistry and microbial ecology in alkaline hot springs of Ambitle Island, Papua New Guinea. Extremophiles 18:763–778. https://doi.org/10.1007/s00792-014-0657-6
Podar PT, Yang Z, Björnsdóttir SH, Podar M (2020) Comparative analysis of microbial diversity across temperature gradients in hot springs from Yellowstone and Iceland. Front Microbiol 11:625. https://doi.org/10.3389/fmicb.2020.01625
García-García N, Tamames J, Linz AM, Pedrós-Alió C, Puente-Sánchez F (2019) Microdiversity ensures the maintenance of functional microbial communities under changing environmental conditions. ISME J 13:2969–2983. https://doi.org/10.1038/s41396-019-0487-8
Overmann J, Abt B, Sikorski J (2017) Present and future of culturing bacteria. Annu Rev Microbiol 71:711–730. https://doi.org/10.1146/annurev-micro-090816-093449
Handelsman J (2004) Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev 68:669–685. https://doi.org/10.1128/MMBR.68.4.669-685.2004
Sharon I, Banfield JF (2013) Genomes from metagenomics. Science 342:1057–1058. https://doi.org/10.1126/science.1247023
Swingley WD, Meyer-Dombard DR, Shock EL, Alsop EB, Falenski HD, Havig JR et al (2012) Coordinating environmental genomics and geochemistry reveals metabolic transitions in a hot spring ecosystem. PLoS ONE 7:e38108. https://doi.org/10.1371/journal.pone.0038108
Satoh T, Watanabe K, Yamamoto H, Yamamoto S, Kurosawa N (2013) Archaeal community structures in the solfataric acidic hot springs with different temperatures and elemental compositions. Archaea 2013:723871. https://doi.org/10.1155/2013/723871
Inskeep WP, Rusch DB, Jay ZJ, Herrgard MJ, Kozubal MA et al (2010) Metagenomes from high temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS ONE 5:e9773. https://doi.org/10.1371/journal.pone.0009773
Wemheuer B, Taube R, Akyol P, Wemheuer F, Daniel R (2013) Microbial diversity and biochemical potential encoded by thermal spring metagenomes derived from the Kamchatka Peninsula. Archaea 13:136714. https://doi.org/10.1155/2013/136714
Delgado-Serrano L, López G, Bohorquez LC, Bustos JR, Rubiano C et al (2014) Neotropical Andes hot springs harbor diverse and distinct planktonic microbial communities. FEMS Microbiol Ecol 89:56–66. https://doi.org/10.1111/1574-6941.12333
Poddar A, Das SK (2018) Microbiological studies of hot springs in India. Rev Arch Microbiol 200:1–18. https://doi.org/10.1007/s00203-017-1429-3
Deep K, Poddar A, Das SK (2013) Anoxybacillus suryakundensis sp. nov, a moderately thermophilic, alkalitolerant bacterium isolated from hot spring at Jharkhand. India. PLoS One 8:e85493. https://doi.org/10.1371/journal.pone.0085493
Poddar A, Lepcha RT, Das SK (2014) Taxonomic study of the genus Tepidiphilus: transfer of Petrobacter succinatimandens to the genus Tepidiphilus as Tepidiphilus succinatimandens comb. nov.; emended description of genus Tepidiphilus and description of Tepidiphilus thermophilus sp. nov. isolated from a terrestrial hot spring in India. Int J Syst Evol Microbiol 64:228–235. https://doi.org/10.1099/ijs.0.056424-0
Badhai J, Ghosh TS, Das SK (2015) Taxonomic and functional characteristics of microbial communities and their correlation with physicochemical properties of four geothermal springs in Odisha. India Front Microbiol 6:1166. https://doi.org/10.3389/fmicb.2015.01166
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Chen S, Zhou Y, Chen Y, Gu J (2018) Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. https://doi.org/10.1093/bioinformatics/bty560
Li D, Liu CM, Luo R, Sadakane K, Lam TW (2015) MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. https://doi.org/10.1093/bioinformatics/btv033
Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW et al (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. https://doi.org/10.1186/1471-2105-11-119
Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Method 12(59):60. https://doi.org/10.1038/nmeth.3176
Huson DH, Beier S, Flade I, Górska A, El-Hadidi M et al (2016) MEGAN community edition—interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput Biol. https://doi.org/10.1371/journal.pcbi.1004957
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. https://doi.org/10.1093/nar/28.1.27
Hsieh TC, Ma KH, Chao A (2016) INEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol Evol 7:1451–1456. https://doi.org/10.1111/2041-210X.12613
McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. https://doi.org/10.1371/journal.pone.0061217
Wickham H (2011) ggplot2. Wiley Interdiscip Rev Comput Stat. https://doi.org/10.1002/wics.147
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. https://doi.org/10.1128/AEM.71.12.8228-8235.2005
Suzuki R, Shimodaira H (2006) Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22:1540–1542. https://doi.org/10.1093/bioinformatics/btl117
Pagaling E, Wang H, Venables M, Wallace A, Grant WD et al (2009) Microbial biogeography of six salt lakes in inner Mongolia, China, and a salt lake in Argentina. Appl Environ Microbiol 75:5750–5760. https://doi.org/10.1128/AEM.00040-09
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics of software package for education and data analysis. Palaeontol Electron 4(9):18
Parks DH, Tyson GW, Hugenholtz P, Beiko RG (2014) STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30(3123):3124. https://doi.org/10.1093/bioinformatics/btu494
Woese CR, Kandler O, Wheelis ML (1990) Towards a natural system of organisms: proposal for the domains archaea, bacteria, and eucarya. Proc Natl Acad Sci U S A 87:4576–4579. https://doi.org/10.1073/pnas.87.12.4576
Stetter KO (2006) Hyperthermophiles in the history of life. Phil Trans R Soc B Biol Sci 361:1837–1843. https://doi.org/10.1098/rstb.2006.1907
Tang J, Liang Y, Jiang D, Li L, Luo Y et al (2018) Temperature-controlled thermophilic bacterial communities in hot springs of western Sichuan. China BMC Microbiol 18:134. https://doi.org/10.1186/s12866-018-1271-z
Esposito A, Engel M, Ciccazzo S, Daprà L, Penna D et al (2016) Spatial and temporal variability of bacterial communities in high alpine water spring sediments. Res Microbiol 167:325–333. https://doi.org/10.1016/j.resmic.2015.12.006
Colman DR, Lindsay MR, Amenabar MJ, Boyd ES (2019) The intersection of geology, geochemistry, and microbiology in continental hydrothermal systems. Astrobiology 19:1505–1522. https://doi.org/10.1089/ast.2018.2016
Zeng Y, Feng F, Medová H, Dean J, Koblížek M (2014) Functional type 2 photosynthetic reaction centers found in the rare bacterial phylum gemmatimonadetes. Proc Natl Acad Sci USA 111:7795–7800. https://doi.org/10.1073/pnas.1400295111
Sharp CE, Brady GH, Sharp SE, Grasby MB, Stott M et al (2014) Humboldt’s spa: microbial diversity is controlled by temperature in geothermal environments. ISME J 8:1166–1174. https://doi.org/10.1038/ismej.2013.237
Vick TJ, Dodsworth JA, Costa KC, Shock EL, Hedlund BP (2010) Microbiology and geochemistry of Little Hot Creek, a hot spring environment in the Long Valley Caldera. Geobiology 8:140–154. https://doi.org/10.1111/j.1472-4669.2009.00228.x
Wang S, Hou W, Dong H, Jiang H, Huang L et al (2013) Control of temperature on microbial community structure in hot springs of the Tibetan Plateau. PLoS ONE 8:e62901. https://doi.org/10.1371/journal.pone.0062901
Tobler DJ, Benning LG (2011) Bacterial diversity in five Icelandic geothermal waters: temperature and sinter growth rate effects. Extremophiles 15:473–485. https://doi.org/10.1007/s00792-011-0378-z
Cole JK, Peacock JA, Dodsworth AJ, Williams DB, Thompson H et al (2013) Sediment microbial communities in great boiling spring are controlled by temperature and distinct from water communities. ISME J 7:718–729. https://doi.org/10.1038/ismej.2012.157
Cuecas A, Portillo MC, Kanoksilapatham M, Gonzalez JM (2014) Bacterial distribution along a 50 degrees c temperature gradient reveals a parceled out hot spring environment. Microb Ecol 68:729–739. https://doi.org/10.1007/s00248-014-0437-y
Macur RE, Langner HW, Kocar BD, Inskeep WP (2004) Linking geochemical processes with microbial community analysis: successional dynamics in an arsenic-rich, acid-sulphate–chloride geothermal spring. Geobiology 2:163–177. https://doi.org/10.1111/j.1472-4677.2004.00032.x
Bräsen C, Esser D, Rauch B, Siebers B (2014) Carbohydrate metabolism in archaea: current insights into unusual enzymes and pathways and their regulation. Microbiol Mol Biol Rev 78:89–175. https://doi.org/10.1128/MMBR.00041-13
Crain AV, Broderick JB (2014) Pyruvate formate-lyase and its activation by pyruvate formate-lyase activating enzyme. J Biol Chem 289:5723–5729. https://doi.org/10.1074/jbc.M113.496877
Dunn MF, Ramírez-Trujillo JA, Hernández-Lucas I (2009) Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology 155:3166–3175. https://doi.org/10.1099/mic.0.030858-0
Ragsdale SW, Pierce E (2008) Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochim Biophys 1784:1873–1898. https://doi.org/10.1016/j.bbapap.2008.08.012
Sapra R, Bagramyan K, Adams MWW (2003) A simple energy-conserving system: proton reduction coupled to proton translocation. Proc Natl Acad Sci USA 100:7545–7550. https://doi.org/10.1073/pnas.1331436100
Baker BJ, Saw JH, Lind AE, Lazer CS, Hinrichs K-U et al (2016) Genomic inference of the metabolism of cosmopolitan subsurface archaea, hadesarchaea. Nat Microbiol 1:16002. https://doi.org/10.1038/nmicrobiol.2016.2
Kelly DP, Shergill JK, Lu WP, Wood AP (1997) Oxidative metabolism of inorganic sulfur compounds by bacteria. Ant Van Leeuwenhoek 71:95–107. https://doi.org/10.1023/a:1000135707181
Liu Y, Beer LL, Whitman WB (2012) Sulfur metabolism in archaea reveals novel processes. Environ Microbiol 14:2632–2644. https://doi.org/10.1111/j.1462-2920.2012.02783.x
Leig JH (2000) Nitrogen fixation in methanogens: the archaeal perspective. Curr Issues Mol Biol 2:125–131
Gomes ES, Schuch V, de Macedo Lemos EG (2013) Biotechnology of polyketides: new breath of life for the novel antibiotic genetic pathways discovery through metagenomics. Braz J Microbiol 44:1007–1034. https://doi.org/10.1590/s1517-83822013000400002
Acknowledgements
The author T.D. acknowledges the Department of Biotechnology, New Delhi, Government of India for providing the research fellowship and S.D. acknowledges the Institute of Life Sciences, Bhubaneswar for providing the fellowship. We acknowledge the Distributed Information Sub-Center (DISC) at Institute of Life Sciences, Bhubaneswar for the computational facility. The geographical map was created from Google Maps using the Geoplaner software available at www.geoplaner.com.
Funding
This work was supported by the funding received from the Department of Biotechnology, Government of India (D.O.No. BT/BI/04/058/2002 VOL-II) to SKD.
Author information
Authors and Affiliations
Contributions
SKD: developed the concept and designed the experiments. TD and SD: participated in laboratory experiments. TD and SKD: interpreted the data and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethical Approval
Ethical approval was not required to carry out this research work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Debnath, T., Deb, S. & Das, S.K. Influence of Geochemistry in the Tropical Hot Springs on Microbial Community Structure and Function. Curr Microbiol 80, 4 (2023). https://doi.org/10.1007/s00284-022-03118-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03118-7