Abstract
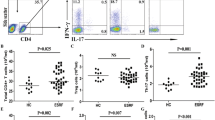
Cytomegalovirus (CMV) reactivation remains a common opportunistic infection with a prominent role in immune reconstitution in organ transplant recipients. CMVs as important drivers of natural killer (NK) cell differentiation has been indicated to prompt several phenotypic and functional alteration in these cells. We aimed to monitor the reconstitution of NK cells and change the signature of inflammatory proteins at the critical phase of CMV reactivation over six months after kidney transplantation. The present study indicated that CMV reactivation is associated with the development of IL-6, IL-10, and cytotoxic granules, including granzyme-B and granulysin, and the drop in the frequency of CD16 + NKG2A-CD57 + NK cell subset in kidney transplant recipients (KTRs) with reactivation versus non- reactivated ones. Our findings describe distinct immune signatures that emerged with CMV reactivation after kidney transplantation, which may be helpful in the timely management of CMV infection in KTRs.




Similar content being viewed by others
References
Karadkhele G, Hogan J, Magua W, Zhang W, Badell IR, Mehta A et al (2021) CMV high-risk status and post-transplant outcomes in kidney transplant recipients treated with Belatacept. Am J Transplant 21(1):208–221. https://doi.org/10.1111/ajt.16132
Jarque M, Crespo E, Melilli E, Gutiérrez A, Moreso F, Guirado L et al (2020) Cellular immunity to predict the risk of cytomegalovirus infection in kidney transplantation: a prospective, interventional, multicenter clinical trial. Clin Infect Dis 71(9):2375–2385. https://doi.org/10.1093/cid/ciz1209
Soleimanian S, Yaghobi R, Karimi MH, Geramizadeh B, Roozbeh J (2022) Loss of CCR7 expression on CD57+ CD56/CD16+ NK cells correlates with viral load in CMV reactivated kidney transplant recipients. Iran J Kidney Dis 1(1):52–62
Martín-Gandul C, Pérez-Romero P, Sánchez M, Bernal G, Suárez G, Sobrino M et al (2013) Determination, validation and standardization of a CMV DNA cut-off value in plasma for preemptive treatment of CMV infection in solid organ transplant recipients at lower risk for CMV infection. J J Clin Virol 56(1):13–18. https://doi.org/10.1016/j.jcv.2012.09.017
Bolovan-Fritts CA, Trout RN, Spector SA (2007) High T-cell response to human cytomegalovirus induces chemokine-mediated endothelial cell damage. Blood 110(6):1857–1863. https://doi.org/10.1182/blood-2007-03-078881
Soleimanian S, Yaghobi R, Karimi MH, Geramizadeh B, Roozbeh J, Hossein Aghdaie M et al (2021) Circulating NKG2C + NK cell expressing CD107a/LAMP-1 subsets at the onset of CMV reactivation in seropositive kidney transplant recipients. Transpl Immunol 69:101460. https://doi.org/10.1016/j.trim.2021.101460
Mena-Romo JD, Pérez Romero P, Martín-Gandul C, Gentil M, Suárez-Artacho G, Lage E et al (2017) CMV-specific T-cell immunity in solid organ transplant recipients at low risk of CMV infection Chronology and applicability in preemptive therapy. J Infect 75(4):336–345. https://doi.org/10.1016/j.jinf.2017.05.020
Hall VG, Humar A, Kumar D (2022) Utility of Cytomegalovirus Cell-Mediated Immunity Assays in Solid Organ Transplantation. J Clin Microbiol 60(8):e0171621. https://doi.org/10.1128/jcm.01716-21
Farzamikia N, Hejazian SM, Haghi M, Hejazian SS, Zununi Vahed S, Ardalan M (2022) Evaluation of telomeric KIR genes and their association with CMV infection in kidney transplant recipients. Immunogenetics 74(2):207–212. https://doi.org/10.1007/s00251-021-01245-2
Kuijpers TW, Baars PA, Dantin C, van den Burg M, van Lier RA, Roosnek E (2008) Human NK cells can control CMV infection in the absence of T cells. Blood 112(3):914–915. https://doi.org/10.1182/blood-2008-05-157354
De Maria A, Bozzano F, Cantoni C, Moretta L (2011) Revisiting human natural killer cell subset function revealed cytolytic CD56dimCD16+ NK cells as rapid producers of abundant IFN-γ on activation. Proc Natl Acad Sci U S A 108(2):728–732. https://doi.org/10.1073/pnas.1012356108
Gumá M, Angulo A, Vilches C, Gómez-Lozano N, Malats N, López-Botet M (2004) Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 104(12):3664–3671. https://doi.org/10.1182/blood-2004-05-2058
DeWolfe D, Aid M, McGann K, Ghofrani J, Geiger E, Helzer C et al (2019) NK cell contributes to the immune risk profile in kidney transplant candidates. Front Immunol 23(10):1890. https://doi.org/10.3389/fimmu.2019.01890
Dendle C, Gan PY, Polkinghorne KR, Ngui J, Stuart RL, Kanellis J et al (2019) Natural killer cell function predicts severe infection in kidney transplant recipients. Am J Transplant 19(1):166–177. https://doi.org/10.1111/ajt.14900
Poli A, Michel T, Thérésine M, Andrès E, Hentges F, Zimmer J (2009) CD56bright natural killer (NK) cells: an important NK cell subset. Immunology 126(4):458–465. https://doi.org/10.1111/j.1365-2567.2008.03027.x
Mavilio D, Lombardo G, Benjamin J, Kim D, Follman D, Marcenaro E et al (2005) Characterization of CD56–/CD16+ natural killer (NK) cells: a highly dysfunctional NK subset expanded in HIV-infected viremic individuals. Proc Natl Acad Sci U S A 102(8):2886–2891. https://doi.org/10.1073/pnas.0409872102
Kärre K (2008) Natural killer cell recognition of missing self. Nat Immunol 9(5):477–480. https://doi.org/10.1038/ni0508-477
Bhatnagar N, Ahmad F, Hong HS, Eberhard J, Lu IN, Ballmaier M et al (2014) FcγRIII (CD16)-mediated ADCC by NK cells is regulated by monocytes and FcγRII (CD32). Eur J Immunol 44(11):3368–3379. https://doi.org/10.1002/eji.201444515
Sivori S, Vacca P, Del Zotto G, Munari E, Mingari MC, Moretta L (2019) Human NK cells: surface receptors, inhibitory checkpoints, and translational applications. Cell Mol Immunol 16(5):430–441. https://doi.org/10.1038/s41423-019-0206-4
Yaghobi R, Behzad-Behbahani A, Sabahi F, Roustaee MH, Alborzi A, Ramzi M, Nourani H (2005) Comparative analysis of a double primer PCR assay with plasma, leukocytes and antigenemia for diagnosis of active human cytomegalovirus infection in bone marrow transplant patients. Bone Marrow Transplant 35(6):595–599. https://doi.org/10.1038/sj.bmt.1704797
Afshari A, Yaghobi R, Karimi MH, Darbouy M, Azarpira N, Geramizadeh B et al (2015) IL-17 mRNA expression and cytomegalovirus infection in liver transplant patients. Exp Clin Transplant 13(Suppl 1):83–89
Pazina T, MacFarlane AWt, Bernabei L, Dulaimi E, Kotcher R, Yam C et al (2021) Alterations of NK cell phenotype in the disease course of multiple myeloma. Cancers (Basel) 13(2):226. https://doi.org/10.3390/cancers13020226
Carrillo-Bustamante P, Keşmir C, de Boer RJ (2016) The evolution of natural killer cell receptors. Immunogenetics 68(1):3–18. https://doi.org/10.1007/s00251-015-0869-7
Soleimanian S, Yaghobi R (2020) Harnessing memory NK cell to protect against COVID-19. Front Pharmacol 20(11):1309. https://doi.org/10.3389/fphar.2020.01309
Foley B, Cooley S, Verneris MR, Curtsinger J, Luo X, Waller EK et al (2012) Human cytomegalovirus (CMV)-induced memory-like NKG2C+ NK cells are transplantable and expand in vivo in response to recipient CMV antigen. J Immunol 189(10):5082–5088. https://doi.org/10.4049/jimmunol.1201964
Young A, Ngiow SF, Gao Y, Patch A-M, Barkauskas DS, Messaoudene M et al (2018) A2AR adenosine signaling suppresses natural killer cell maturation in the tumor microenvironment. C Cancer Res 78(4):1003–1016. https://doi.org/10.1158/0008-5472.CAN-17-2826
Bozzano F, Della Chiesa M, Pelosi A, Antonini F, Ascierto ML, Del Zotto G et al (2021) HCMV-controlling NKG2C+ NK cells originate from novel circulating inflammatory precursors. J Allergy Clin Immunol 147(6):2343–2357. https://doi.org/10.1016/j.jaci.2020.12.648
Jang JE, Hwang DY, Chung H, Kim S-J, Eom J-I, Jeung H-K et al (2019) Early cytomegalovirus reactivation and expansion of CD56brightCD16dim/− DNAM1+ natural killer cells are associated with antileukemia effect after haploidentical stem cell transplantation in acute leukemia. Biol Blood Marrow Transplant 25(10):2070–2078. https://doi.org/10.1016/j.bbmt.2019.06.008
Rizzo R, Zatelli MC, Rotola A, Cassai E, Degli Uberti E, Di Luca D et al (2016) Increase in peripheral CD3− CD56 bright CD16− natural killer cells in hashimoto’s thyroiditis associated with HHV-6 infection. Adv Exp Med Biol 897:113–120. https://doi.org/10.1007/5584_2015_5010
Oliviero B, Mantovani S, Varchetta S, Mele D, Grossi G, Ludovisi S et al (2017) Hepatitis C virus-induced NK cell activation causes metzincin-mediated CD16 cleavage and impaired antibody-dependent cytotoxicity. J Hepatol 66(6):1130–1137. https://doi.org/10.1016/j.jhep.2017.01.032
Zimmer J, Bausinger H, Andrès E, Donato L, Hanau D, Hentges F et al (2007) Phenotypic studies of natural killer cell subsets in human transporter associated with antigen processing deficiency. PLoS ONE 2(10):e1033. https://doi.org/10.1371/journal.pone.0001033
Bielekova B, Catalfamo M, Reichert-Scrivner S, Packer A, Cerna M, Waldmann TA et al (2006) Regulatory CD56bright natural killer cells mediate immunomodulatory effects of IL-2Rα-targeted therapy (daclizumab) in multiple sclerosis. Proc Natl Acad Sci U S A 103(15):5941–5946. https://doi.org/10.1073/pnas.0601335103
Nielsen CM, White MJ, Goodier MR, Riley EM (2013) Functional significance of CD57 expression on human NK cells and relevance to disease. Front Immunol 9(4):422. https://doi.org/10.3389/fimmu.2013.00422
Chan A, Hong DL, Atzberger A, Kollnberger S, Filer AD, Buckley CD et al (2007) CD56bright human NK cells differentiate into CD56dim cells: role of contact with peripheral fibroblasts. J Immunol 179(1):89–94. https://doi.org/10.4049/jimmunol.179.1.89
Lopez-Vergès S, Milush JM, Pandey S, York VA, Arakawa-Hoyt J, Pircher H et al (2010) CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood, J Am Soc Hematol 116(19):3865–3874
Björkström NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA et al (2010) Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood 116(19):3853–3864. https://doi.org/10.1182/blood-2010-04-281675
Ali TH, Pisanti S, Ciaglia E, Mortarini R, Anichini A, Garofalo C et al (2014) Enrichment of CD56 dim KIR+ CD57+ highly cytotoxic NK cells in tumour-infiltrated lymph nodes of melanoma patients. Nat Commun 4(5):5639. https://doi.org/10.1038/ncomms6639
Soleimanian S, Yaghobi R, Karimi MH, Geramizadeh B, Roozbeh J, Aghdaie MH et al (2021) Circulating NKG2C+ NK cell expressing CD107a/LAMP-1 subsets at the onset of CMV reactivation in seropositive kidney transplant recipients. Transpl Immunol 69:101460. https://doi.org/10.1016/j.trim.2021.101460
Soleimanian S, Yaghobi R, Karimi MH, Geramizadeh B, Roozbeh J (2021) The direct influence of cytomegalovirus lysate on the natural killer cell receptor repertoire. Iran J Allergy Asthma Immunol 20(6):721–733. https://doi.org/10.18502/ijaai.v20i6.8023
Clark SE, Burrack KS, Jameson SC, Hamilton SE, Lenz LL (2019) NK cell IL-10 production requires IL-15 and IL-10 driven STAT3 activation. Front Immunol 4(10):2087. https://doi.org/10.3389/fimmu.2019.02087
Chan YLT, Zuo J, Inman C, Croft W, Begum J, Croudace J et al (2018) NK cells produce high levels of IL-10 early after allogeneic stem cell transplantation and suppress development of acute GVHD. Eur J Immunol 48(2):316–329. https://doi.org/10.1002/eji.201747134
Stacey MA, Marsden M, Wang EC, Wilkinson GW, Humphreys IR (2011) IL-10 restricts activation-induced death of NK cells during acute murine cytomegalovirus infection. J Immunol 187(6):2944–2952. https://doi.org/10.4049/jimmunol.1101021
Alter G, Malenfant JM, Altfeld M (2004) CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods 294(1–2):15–22. https://doi.org/10.1016/j.jim.2004.08.008
Rabe T, Lazar K, Cambronero C, Goelz R, Hamprecht K (2020) Human cytomegalovirus (HCMV) reactivation in the mammary gland induces a proinflammatory cytokine shift in breast milk. Microorganisms 8(2):289. https://doi.org/10.3390/microorganisms8020289
Holder KA, Grant MD (2019) Human cytomegalovirus IL-10 augments NK cell cytotoxicity. J Leukoc Biol 106(2):447–454. https://doi.org/10.1002/JLB.2AB0418-158RR
Nowacki TM, Bettenworth D, Ross M, Heidemann J, Lehmann PV, Lügering A (2012) Cytomegalovirus (CMV)-specific perforin and granzyme B ELISPOT assays detect reactivation of CMV infection in inflammatory bowel disease. Cells 1(2):35–50
Wever PC, Spaeny LH, van der Vliet HJ, Rentenaar RJ, Wolbink AM, Surachno J et al (1999) Expression of granzyme‐B during primary cytomegalovirus infection after renal transplantation. J Infect Dis 179(3):693–696. https://doi.org/10.1086/314629
Krensky A, Clayberger C (2009) Biology and clinical relevance of granulysin. Tissue Antigens 73(3):193–198. https://doi.org/10.1111/j.1399-0039.2008.01218.x
Sarwal MM, Jani A, Chang S, Huie P, Wang Z, Salvatierra O Jr et al (2001) Granulysin expression is a marker for acute rejection and steroid resistance in human renal transplantation. Hum Immunol 62(1):21–31. https://doi.org/10.1016/s0198-8859(00)00228-7
Fauriat C, Long EO, Ljunggren H-G, Bryceson YT (2010) Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood 115(11):2167–2176. https://doi.org/10.1182/blood-2009-08-238469
Froeschle GM, Bedke T, Boettcher M, Huber S, Singer D, Ebenebe CU (2021) T cell cytokines in the diagnostic of early-onset sepsis. Pediatr Res 90(1):191–196. https://doi.org/10.1038/s41390-020-01248-x
Acknowledgements
This work was supported by Shiraz University of Medical Sciences [Grant No.13843]. We thank the Abu Ali Sina Transplant Hospital, Shiraz (Shiraz, Iran), and participants for collecting samples.
Author information
Authors and Affiliations
Contributions
SS, RY, MH-K, BG, and JR conceived the idea and designed the study. SS has performed the experiments and written the manuscript. RY, MH-K, BG, and JR critically revised the article. All authors contributed to the article and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no financial conflicts of interest.
Ethical Approval
The studies involving human participants were reviewed and approved by the local Ethics Committee of Shiraz University of Medical Sciences (IR.SUMS.REC. 1396. S289).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Soleimanian, S., Yaghobi, R., Karimi, M.H. et al. Altered Signatures of Plasma Inflammatory Proteins and Phonotypic Markers of NK Cells in Kidney Transplant Patients upon CMV Reactivation. Curr Microbiol 80, 9 (2023). https://doi.org/10.1007/s00284-022-03116-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03116-9