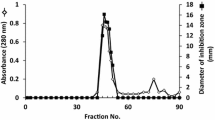
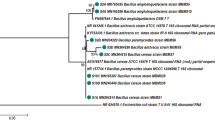
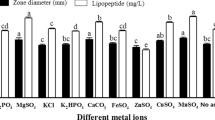
Abstract
A bacterium strain isolated from freshwater sediment of San Pablo river of Santiago de Cuba, Cuba was identified as a Bacillus sp. by Matrix-Assisted Laser Desorption/Ionization Time Of Flight Mass Spectrometry. A 16S rRNA gene analysis showed that the isolate A3 belongs to the operational group Bacillus amyloliquefaciens, while the phylogenetic analysis of the gyrA gene sequence grouped it within B. amyloliquefaciens subsp. plantarum cluster, referred now as Bacillus velezensis. In vitro antibacterial studies demonstrated the capacity of the isolate A3 to produce bioactive metabolites against Bacillus subtilis ATCC 11,778, Bacillus cereus ATCC 6633, and Staphylococcus aureus ATCC 25,923 by cross-streak, overlay, and microdilution methods. The strain also showed a high potential against the multidrug-resistant Staphylococcus aureus ATCC 700,699, ATCC 29,213, and ATCC 6538. At pH 8 and 96 h in the medium 2 of A3 culture conditions, the produced metabolites with antibacterial potential were enhanced. Some alterations in the morphology of the phytopathogens Aspergillus niger ATCC 9642, Alternaria alternata CECT 2662, and Fusarium solani CCEBI 3094 were induced by the cell-free supernatant of B. velezensis A3. A preliminary study of the nature of the bioactive compounds produced by the strain A3 showed the presence of both lipids and peptides in the culture. Those results highlight B. velezensis A3 as a promissory bacterium capable to produce bioactive metabolites with antibacterial and antifungal properties against pathogens.





Similar content being viewed by others
Data Availability
All data generated during this study are included in this article and its supplementary information file.
Code Availability
Not applicable.
References
Zhi-Wen Y, Yan-Li Z, Man Y, Wei-Jun F (2015) Clinical treatment of pandrug-resistant bacterial infection consulted by clinical pharmacist. Saudi Pharm J 23:377–380
Mann A, Nehra K, Rana JS, Dahiya T (2021) Antibiotic resistance in agriculture: perspectives on uncoming strategies to overcome upsurge in resistance. Sciences 2:10030
Viswanathan VK (2014) Off-label abuse of antibiotics by bacteria. Gut Microbes 5:3–4
Shafi J, Tian H, Ji M (2017) Bacillus species as versatile weapons for plant pathogens: a review. Biotechnol Biotech Equip 31:446–459
Eon JS, Choi HS (2016) Inhibition of Bacillus cereus growth and toxin production by Bacillus amyloliquefaciens RD7-7 in fermented soybean products. J Microbiol Biotechnol 26:44–55
Lee JY, Shim JM, Yao Z, Liu X, Lee KW, Kim HJ, Ham KS, Kim JH (2016) Antimicrobial activity of Bacillus amyloliquefaciens EMD17 isolated from Cheonggukjang and potential use as a starter for fermented soy foods. Food Sci Biotechnol 30(25):525–532
Singhal N, Kumar M, Kanaujia PK, Virdi JS (2015) MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol 6:791
AlMasoud N, Xu Y, Nicolaou N, Goodacre R (2014) optimization of matrix assisted laser desorption/ionization time of fight mass spectrometry (MALDI-TOF-MS) for the characterization of Bacillus and Brevibacillus species. Anal Chim Acta 840:49–57
Ha M, Jo HJ, Choi EK, Kim Y, Kim J, Cho HJ (2019) Reliable identification of Bacillus cereus group species using low mass biomarkers by MALDI-TOF MS. J Microbiol Biotechnol 29:887–896
Cuénod A, Foucault F, Pflüger V, Egli A (2021) Factors associated with MALDI-TOF mass spectral quality of species identification in clinical routine diagnostics. Front Cell Infect Microbiol 11:646648
Rooney AP, Price NP, Ehrhardt C, Swezey JL, Bannan JD (2009) Phylogeny and molecular taxonomy of the Bacillus subtilis species complex and description of Bacillus subtilis subsp. inaquosorum subsp. nov. Int J Syst Evol Microbiol 59:2420–2436
Vandamme P (2007) Taxonomy and classification of bacteria. In: Murray PR, Baron EJ, Jorgenson JH, Landry ML, Pfaller MA (eds) Manual of clinical microbiology, 9th edn. DC, USA, Washington, pp 275–290
Bauer AW, Kirby WMM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 36:493–496 (PMID: 5325707)
Tagg JR, McGiven AR (1971) Assay system for bacteriocins. Appl Microbiol 21:943–943
Méndez D, Escalona-Arranz J, Foubert K, Matheeussen A, Van der Auwera A, Piazza S, Cuypers A, Cos P, Pieters L (2021) Chemical and pharmacological potential of Coccoloba cowellii, an endemic endangered plant from Cuba. Molecules 26(4):935
Wieme AD, Spitaels F, Aerts M, De Bruyne K, Landschoot AV, Vandamme P (2014) Effects of growth medium on matrix-assisted laser desorption-ionization time of flight mass spectra: a case study of acetic acid bacteria. Appl Environ Microbiol 80:1528–1538
Niedermeyer THJ, Strohalm M (2012) mMass as a software tool for the annotation of cyclic peptide tandem mass spectra. PLoS ONE 7:e44913
Snauwaert I, Papalexandratou Z, De Vuyst L, Vandamme P (2013) Characterization of strains of Weissella fabalis sp. nov. and Fructobacillus tropaeoli from spontaneous cocoa bean fermentations. Int J Syst Evol Microbiol 63:1709–1716
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Pruesse E, Peplies J, Glöckner FO (2012) SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinform 28:1823–1829
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Chun J, Bae KS (2000) Phylogenetic analysis of Bacillus subtilis and related taxa based on partial gyrA gene sequences. Antonie Van Leeuwenhoek 78:123–127
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Bérdy J (2005) Bioactive microbial metabolites: a personal view. J Antibiot 58:1–26
Huschek D, Witzei K (2019) Rapid dereplication of microbial isolates using matrix-assisted laser desorption ionization time-of-flight mass spectrometry: a mini-review. J Adv Res 19:99–104
Han SS, Jeong YS, Choi SK (2021) Current scenario and challenges in the direct identification of microorganisms using MALDI TOF MS. Microorganisms 9:1917
Dunlap C, Kim SJ, Kwon SW, Rooney A (2016) Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens, Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. plantarum and ‘Bacillus oryzicola’ are later heterotypic synonyms of of Bacillus velezensis based on phylogenomics. Int J Syst Evol Microbiol 66:1212–1217
Toral L, Rodríguez M, Béjar V, Sampedro I (2018) Antifungal activity of lipopeptides from Bacillus XT1 CECT 8661 against Botrytis cinerea. Front Microbiol 9:1315
Li X, Gao X, Zhang S, Jiang Z, Yang H, Liu X, Jiang Q, Zhang X (2020) Characterization of a Bacillus velezensis with antibacterial activity and inhibitory effect on common aquatic pathogens. Aquac 523:735165
Rabbee MF, Baek KH (2020) Antimicrobial activities of lipopeptides and polyketides of Bacillus velezensis for agricultural applications. Molecules 25(21):4973
Baharudin M, Ngalimat MS, Mohd Shariff F, Balia Yusof ZN, Karim M, Baharum SN, Sabri S (2021) Antimicrobial activities of Bacillus velezensis strains isolated from stingless bee products against methicillin-resistant Staphylococcus aureus. PLoS ONE 16:e0251514
Pokhrel R, Bhattarai N, Baral P, Gerstman BS, Park JH, Handfield M, Chapagain PP (2019) Molecular mechanisms of pore formation and membrane disruption by the antimicrobial lantibiotic peptide Mutacin 1140. Phys Chem Chem Phys 21(23):12530–12539
Benfield AH, Henriques ST (2020) Mode-of-action of antimicrobial peptides: membrane disruption vs intracellular mechanisms. Front Med Technol. https://doi.org/10.3389/fmedt.2020.610997
Silva M, Pereira A, Teixeira D, Candeias A, Caldeira AT (2016) Combined use of NMR, LC-ESI-MS and antifungal tests for rapid detection of bioactive lipopeptides produced by Bacillus. Adv Microbiol 6:788–796
Meena KR, Tandon T, Sharma A, Kanwar SS (2018) Lipopeptide antibiotic production by Bacillus velezensis KLP2016. J App Pharm Sci 8(03):091–098
Orberá TM, Serrat MJ, Ortega E (2014) Potential applications of Bacillus subtilis strain SR/B-16 for the control of phytopathogenic fungi in economically relevant crops. Biotechnol Apl 31:7–12
Andrić S, Meyer T, Ongena M (2020) Bacillus responses to plant-associated fungal and bacterial communities. Front Microbiol 11:1350
Grady EN, MacDonald J, Ho MT, Weselowski B, McDowell T, Solomon O, Renaud J (2019) Yuan ZC (2019) Characterization and complete genome analysis of the surfactin-producing, plant-protecting bacterium Bacillus velezensis 9D–6. BMC Microbiol 19:5
Meena KR, Sharma A, Kumar R, Kanwar SS (2020) Two factor at a time approach by response surface methodology to aggrandize the Bacillus subtilis KLP2015 surfactin lipopeptide to use as antifungal agent. J King Saud Univ Sci 32:337–348
Qiu J, Kirsh LE (2014) Evaluation of lipopeptide (Daptomycin) aggregation using fluorescence, light scattering, and nuclear magnetic resonance spectroscopy. J Pharm Sci 103(3):853–861
Hmidet N, Ayed HB, Jacques P, Nasri M (2017) Enhancement of surfactin and fengycin production by Bacillus mojavensis A21: application for diesel biodegradation. BioMed Res Int 3:1–8
Phulpoto IA, Yu Z, Hu B, Wang Y, Ndayisenga F, Li J, Liang H, Qazi MA (2020) Production and characterization of surfactin-like biosurfactant produced by novel strain Bacillus nealsonii S2MT and it’s potential for oil contaminated soil remediation. Microb Cell Fact 19:145
Barale SS, Ghane SG, Sonawane KD (2022) Purification and characterization of antibacterial surfactin isoforms produced by Bacillus velezensis SK. AMB Express 12(1):7
Pournejati R, Karbalaei-Heidari HR (2020) Optimization of fermentation conditions to enhance cytotoxic metabolites production by Bacillus velezensis strain RP137 from the Persian Gulf. Avicenna J Med Biotechnol 12(2):116–123
Jamil B, Hasan F, Hameed A, Ahmed S (2007) Isolation of Bacillus subtilis MH-4 from soil and its potential of polypeptidic antibiotic production. Pak J Pharm Sci 20(1):26–31
Ghribi D, Ellouze-Chaabouni S (2011) Enhancement of Bacillus subtilis lipopeptide biosurfactants production through optimization of medium composition and adequate control of aeration. Biotech Res Int 2011:1–6
Nehal N, Singh P (2021) Optimization of cultural condition of Bacillus sp. MZ540316: improve biodegradation efficiency of lipopeptide biosurfactant against polyethylene. Biomass Conv Bioref. https://doi.org/10.1007/s13399-021-02042-3
Acknowledgements
The authors would like to thanks to the BCCM/LMG Bacterial Collection regarding bacterial identification. We appreciate the help of Prof. dr. Paul Cos (Department of Pharmaceutical Sciences, University of Antwerp, Belgium), and Eliza Depoorter (Laboratory of Microbiology, Faculty of Sciences, Ghent University, Belgium) for the help with antibacterial activity by broth microdilution and overlay methods, respectively.
Funding
This work was supported by the VLIR-UOS (Flemish Interuniversity Council-University Cooperation for Development) in the context of the Institutional University Cooperation Program with Universidad de Oriente; especially, by means of the P-3 project “Natural Products and Pharmaceutical Services to improve the patient quality of life in Eastern Cuban Hospital’s,” The BCCM/LMG Bacteria Collection is supported by the Federal Public Planning Service—Science Policy, Belgium.
Author information
Authors and Affiliations
Contributions
MIC Conceptualization, Performing experiments, Analysis, and Interpretation of data, Writing—Original Draft; SR & GLL Conceptualization, Writing—Review & Editing; PV Resources, Critical revision of the article; AW Resources, Analysis, and interpretation of data; AF Analysis and Interpretation of data; JG, DR, JB, YL, LP, & TLM Performing experiments. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical Approval
This article does not contain any studies with human participants or animals.
Consent for Publication
Not applicable.
Consent to Participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Camacho, M.I., García, J.M., Roget, D. et al. Isolation and Identification of a Bacillus sp. from Freshwater Sediment Displaying Potent Activity Against Bacteria and Phytopathogen Fungi. Curr Microbiol 79, 398 (2022). https://doi.org/10.1007/s00284-022-03090-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03090-2