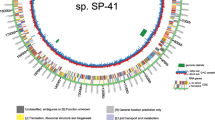
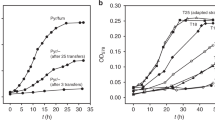
Abstract
The Hyf-type formate hydrogen lyase (FHL) complex was first proposed based on sequence comparisons in Escherichia coli in 1997 (Andrews et al. in Microbiology 143:3633–3647, 1997). The hydrogenase in the Hyf-type FHL was estimated to be a proton-translocating energy-conserving [NiFe]-hydrogenase. Although the structure of FHL is similar to that of complex I, silent gene expression in E. coli has caused delays in unveiling the genetic and biochemical features of the FHL. The entire set of genes required for Hyf-type FHL synthesis has also been found in the genome sequences of Vibrio tritonius in 2015 (Matsumura et al. in Int J Hydrog Energy 40:9137–9146, 2015), which produces more hydrogen (H2) than E. coli. Here we investigate the physiological characteristics, genome comparisons, and gene expressions to elucidate the genetic backgrounds of Hyf-type FHL, and how Hyf-type FHL correlates with the higher H2 production of V. tritonius. Physiological comparisons among the seven H2-producing vibrios reveal that V. porteresiae and V. tritonius, grouped in the Porteresiae clade, show greater capacity for H2 production than the other species. The structures of FHL-Hyp gene clusters were closely related in both Porteresiae species, but differed from those of the other species with the presence of hupE, a possible nickel permease gene. Interestingly, deeper genome comparisons revealed the co-presence of nickel ABC transporter genes (nik) with the Hyf-type FHL gene only on the genome of the Porteresiae clade species. Therefore, active primary Ni transport might be one of the key factors characterizing higher H2 production in V. tritonius. Furthermore, the expression of FHL gene cluster was significantly up-regulated in V. tritonius cells stimulated with formate, indicating that formate is likely to be a control factor for the gene expression of V. tritonius FHL in a similar way to the formate regulon encoding the E. coli FHL.



Similar content being viewed by others
Data Availability
RNA-Seq reads were deposited DBJ/GenBank/ENA under accession number DRA014595.
Code Availability
Not applicable.
References
Andrews SC, Berks BC, McClay J, Ambler A, Quail MA, Golby P, Guest JR (1997) A 12-cistron Escherichia coli operon (hyf) encoding a putative proton-translocating formate hydrogenlyase system. Microbiology 143:3633–3647
Matsumura Y, Al-saari H, Mino M, Nakagawa S, Maruyama F, Ogura Y, Hayashi T, Kurokawa K, Sawabe T, Sawabe T (2015) Identification of a gene cluster responsible for hydrogen evolution in Vibrio tritonius strain AM2 with transcriptional analyses. Int J Hydrog Energy 40:9137–9146
Thompson FL, Gevers D, Thompson CC, Dawyndt P, Hoste B, Munn CB (2005) Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl Environ Microbiol 71:5107–5115
Sawabe T, Kita-Tsukamoto K, Thompson FL (2007) Inferring the evolutionary history of vibrios by means of multilocus sequence analysis. J Bacteriol 189:7932–7936
Sawabe T, Ogura Y, Matsumura Y et al (2013) Updating the vibrio clades defined by multilocus sequence phylogeny: proposal of eight new clades, and the description of Vibrio tritonius sp. Nov, Front Microbiol. https://doi.org/10.3389/fmicb.2013.00414
Jiang C, Tanaka M, Nishikawa S, Mino S, Romalde JL, Thompson FL, Gomez-Gil B, Sawabe T (2022) Vibrio clade 3.0: new Vibrionaceae evolutionary units using genome-based approach. Cur Microbiol. https://doi.org/10.1007/s00284-021-02725-0
Gomez-Gil B, Thompson CC, Vicente A, Matsumura Y, Sawabe T, Iida T, Christen R, Sawabe T (2014) Famlily Vibrionaceae. In: Rosenberg E et al (eds) The Prokaryotes, 4th edn. Springer, New York, pp 659–747
Lee JV, Shread P, Furniss AL, Bryant TN (1981) Taxonomy and description of Vibrio fluvialis sp. nov. (Synonym Group F Vibrios, Group EF6). J Appl Bacteriol 50:73–94
Brenner DJ, Hickman-Brenner FW, Lee JV, Steigerwalt AG, Fanning GR, Hollis DG et al (1983) Vibrio furnissii (formerly aerogenic biogroup of Vibrio fluvialis), a new species isolated from human feces and the environment. J Clin Microbiol 18:816–824
Harwood CS (1978) Beneckea gazogenes sp. nov., a red, facultatively anaerobic marine bacterium. Curr Microbiol 1:233–238
Baumann P, Furniss AL, Lee JV (1984) Genus I. Vibrio Pacini 1854, 411 AL. In: Krieg NR (ed) Bergey’s Manual of Systematic Bacteriology, vol 1. Williams & Wilkins, Baltimore, pp 518–538
Shieh WY, Chen AL, Chiu HH (2000) Vibrio aerogenes sp. nov., a facultatively anaerobic marine bacterium that ferments glucose with gas production. Int J Syst Evol Microbiol 50:321–329
Shieh WY, Chen YW, Chaw SM, Chiu HH (2003) Vibrio ruber sp. nov., a red, facultatively anaerobic, marine bacterium isolated from sea water. Int J Syst Evol Microbiol 53:479–484
Kumar NR, Sudha N (2007) Vibrio rhizosphaerae sp. nov., a red-pigmented bacterium that antagonizes phytopathogenic bacteria. Int J Syst Evol Microbiol 57:2241–2246
Pujalte MJ, Ortigosa M, Urdaci MC, Garay E, Grimont PAD (1993) Vibrio mytili sp. nov., from mussels. Int J Syst Bacteriol 43:358–362
Tanaka M, Endo S, Kotake F, Al-Saari N, Amin AKMR, Gao F et al (2017) Vibrio aphrogenes sp. Nov.,in the Rumoiensis clade isolated from a seaweed. PLoS ONE 12(6):e0180053–e0180056
Rameshkumar N, Fukui Y, Sawabe T, Nair S (2008) Vibrio porteresiae sp. nov., a diazotrophic bacterium isolated from a mangrove-associated wild rice (Porteresia coarctata Tateoka). Int J Syst Evol Microbiol 58:1608–1615
Matsumura Y, Sato K, Al-saari H, Nakagawa S, Sawabe T (2014) Enhanced hydrogen production by a newly described heterotrophic marine bacterium, Vibrio tritonius strain AM2, using seaweed as the feedstock. Int J Hydrog Energy 39:7270–7277
Vignais PM, Colbeau A (2004) Molecular biology of microbial hydrogenases. Mol Biol 6:159–188
Nicolet Y, Lemon BJ, Fontecilla-Camps JC, Peters JW (2000) A novel FeS cluster in Fe-only hydrogenases. Trends Biochm Sci 25:138–143
Vignais PM, Billoud B (2007) Occurrence, classification, and biological function of hydrogenases: an overview. Chem Rev 107:4206–4272
Mnatsakanyan N, Bagramyan K, Trchounian A (2004) Hydrogenase 3 but not hydrogenase 4 is major in hydrogen gas production by Escherichia coli formate hydrogenlyase at acidic pH and in the presence of external formate. Cell Biochem Biophys 41:357–366
Noguchi K, Riggins DP, Eldahan KC, Kitko RD, Slonczewski JL (2010) Hydrogenase-3 contributes to anaerobic acid resistance of Escherichia coli. PLoS ONE 5:e10132
Vignais PM, Billoud B, Meyer J (2001) Classification and phylogeny of hydrogenase. FEMS Microbiol Rev 25:455–501
Schmidt O, Wüst PK, Hellmuth S, Borst K, Horn MA, Drake HL (2011) Novel [NiFe]- and [FeFe]-hydrogenase gene transcripts indicative of active facultative aerobes and obligate anaerobes in earthworm gut contents. Appl Environ Microbiol 77:5842–5850
Sawers G (1994) The hydrogenases and formate dehydrogenases of Escherichia coli. Antonie Van Leeuwenhoek 66:57–88
Sawers RG (2005) Formate and its role in hydrogen production in Escherichia coli. Biochem Soc Trans 33:42–46
Casalot L, Rousset M (2001) Maturation of the [NiFe] hydrogenases. Trends Microbiol 9:228–237
Self WT, Hasona A, Shanmugam KT (2004) Expression and regulation of a silent operon, hyf, coding for hydrogenase 4 isoenzyme in Escherichia coli. J Bacteriol 186:580–587
Bagramyan K, Mnatsakanyan N, Poladian A, Vassilian A, Trchounian A (2002) The roles of hydrogenases 3 and 4, and the FoF1-ATPase, in H2 production by Escherichia coli at alkaline and acidic pH. FEBS Lett 516:172–178
Böck A, Sawers G (1996) Fermentation. In: Neidhardt FC (ed) Escherichia coli and Salmonella, cellular and molecular biology, 2nd edn. ASM Press, Washington DC, pp 262–282
Finney AJ, Lowden R, Fleszar M, Albareda M, Coulthurst SJ, Sargent F (2020) The plant pathogen Pectobacterium atrosepticum contains a functional formate hydrogenlyase-2 complex. Mol Microbiol 112:1440–1452
Lindenstrauß U, Pinske C (2019) Dissection of the hydrogen metabolism of the enterobacterium Trabulsiella guamensis: identification of a formate-dependent and essential formate hydrogenlyase complex exhibiting pylogenetic similarity to complex I. J Bacteriol 201:e00160–e219
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T et al (2014) The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Berriman M, Rutherford K (2003) Viewing and annotating sequence data with Artemis. Brief Bioinform 4:124–132
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Thorvaldsdóttir H, Robinson JT, Mesirov JP (2013) Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192
Gyaneshwar P, James EK, Mathan N, Reddy PM, Reinhold-Hurek B, Ladha JK (2001) Endophytic colonization of rice by a diazotrophic strain of Serratia marcescens. J Bacteriol 183:2634–2645
Rowe JL, Starnes GL, Chivers PT (2005) Complex transcriptional control links NikABCDE-dependent nickel transport with hydrogenase expression in Escherichia coli. J Bacteriol 187:6317–6323
Rodionov DA, Hebbeln P, Gelfand MS, Eitinger T (2006) Comparative and functional genomic analysis of prokarytic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J Bacteriol 188:317–327
Remy L, Carrière M, Derré-Bobillot A, Martini C, Sanguinetti M, Borezée-Durant E (2013) The Staphylococcus aureus Opp1 ABC transporter imports nickel and cobalt in zinc-depleted conditions and contributes to virulence. Mol Microbiol 87:730–743
Eitinger T, Mandrand-Berthelot MA (2000) Nickel transport systems in microorganisms. Arch Microbiol 173:1–9
Brito B, Prieto RI, Cabrera E, Mandrand-Berthelot MA, Imperial J, Ruiz-Argüeso T, Palacios JM (2010) Rhizobium leguminosarum hupE encodes a nickel transporter required for hydrogenase activity. J Bacteriol 192:925–935
Rossmann R, Sauter M, Lottspeich F, Böck A (1994) Maturation of the large subunit (HYCE) of Escherichia coli hydrogenase 3 requires nickel incorporation followed by C-terminal processing at Arg537. Eur J Biochem 220:377–384
Moparthi VK, Hägerhäll C (2011) The evolution of respiratory chain complex I from a smaller last common ancestor consisting of 11 protein subunits. J Mol Evol 72:484–497
Marreiros BC, Batista AP, Duarte AM, Pereira MM (2013) A missing link between complex I and group 4 membrane-bound [NiFe] hydrogenases. Biochim Biophys Acta 1827:198–209
McDowall JS, Murphy BJ, Haumann M, Palmer T, Armstrong FA, Sargent F (2014) Bacterial formate hydrogenlyase complex. Proc Natl Acad Sci USA 111:E3948-3956
Baradaran R, Berrisford JM, Minhas GS, Sazanov L (2013) Crystal structure of the entire respiratory complex I. Nature 494:443–448
Barquera B (2014) The sodium pumping NADH:quinone oxidoreductase (Na+-NQR), a unique redox-driven ion pump. J Bioenerg Biomembr 46:289–298
Barquera B, Hellwig P, Zhou W, Morgan JE, Häse CC, Gosink KK et al (2022) Purification and characterization of the recombinant Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry 41:3781–3789
Skibinski DA, Golby P, Chang YS, Sargent F, Hoffman R, Harper R, Guest JR, Attwood MM, Berks BC, Andrews SC (2002) Regulation of the hydrogenase-4 operon of Escherichia coli by the σ54-dependent transcriptional activators FhlA and HyfR. J Bacteriol 184:6642–6653
Kneip C, Lockhart P, Voß C, Maier UG (2007) Nitrogen fixation in eukaryotes—New models for symbiosis. BMC Evol Biol 7:55
Zehr JP, Mellon MT, Zani S (1998) New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl Environ Microbiol 64:3444–3450
Acknowledgements
We would like to thank Professor Suzuki (The Tokyo University), and Mr. Mizukoshi (Hokkaido University) for technical supports.
Funding
This work was supported by the MEXT Kaken 25292122, 16H04976, and 19H03041.
Author information
Authors and Affiliations
Contributions
Conceptualization: TS; Research and Data analysis: YM, KS, and TS; Writing—original draft preparation: YM, KS, and TS; Writing—review and editing: YM, KS, JC, SM, and TS; Resources: TS. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no competing interests.
Ethical Approval
Not applicable.
Consent for Publication
Not applicable.
Consent to Participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
284_2022_3065_MOESM1_ESM.pptx
Supplementary file1 (PPTX 899 kb) Fig. S1. Codon usage of CDSs on FHL-Hyp gene cluster and on chromosome 1 in Vibrio tritonius. All codon including stop codon was shown. The values indicated abundance ratio of each codon. Fig. S2. Schematic of nikABCDE and nikR genes on genomes. The genes responsible for synthesis nickel ABC transporter in genomes of E. coli, V. tritonius, V. porteresiae, and V. aerogenes are shown. Fig. S3. Effect of extracellular formate on H2 production of Vibrio tritonius. (A) Kinetic formate consumption for 96 h of V. tritonius is shown. Error bars represent standard error (N = 3). H2 (B) Kinetic H2 production derived from extracellular formate for 96 h of V. tritonius was shown. Error bars represent standard error (N = 3). Fig. S4. Coverage of RNA-Seq reads mapping to FHL-Hyp gene cluster in Vibrio tritonius. Arrows indicate the predicted discontinuity sites of transcripts of FHL-Hyp gene cluster. Fig. S5. Schematic of nif gene cluster in Vibrio tritonius, Vibrio porteresiae, and Vibrio aerogenes genomes. Table S1. Summary of the genome sequences of seven hydrogen-producing vibrios. Table S2. Genes showing differential expression by mannitol supplementation. Table S3. Genes showing differential expression by formate supplementation.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Matsumura, Y., Sato, K., Jiang, C. et al. Comparative Physiology and Genomics of Hydrogen-Producing Vibrios. Curr Microbiol 79, 360 (2022). https://doi.org/10.1007/s00284-022-03065-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03065-3