Abstract
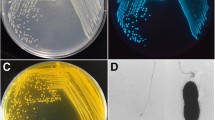
Vibrio parahaemolyticus is a bacterial pathogen in marine aquaculture systems and a major cause of food-borne illnesses worldwide. In the present study, Vibrio phage KIT05 was isolated from water collected from a shrimp farm in the Mekong Delta, Vietnam. It was characterized based on its morphology, growth curve, lytic properties, and genome sequence. Under the electron microscope, KIT05 particles had an icosahedral head with a diameter of 62.3 nm and a short tail of 24.1 nm. The one-step growth curve of KIT05 showed that its latency time was approximately 40 min and burst size was 18 plaque-forming units/cell. The genome of KIT05 comprises 50,628 bp with a GC content of 41.63%. It contains 60 open reading frames that are encoded within both strands and four tRNAs. The presence of direct terminal repeats of 130 bp at both ends of the KIT05 DNA was determined. According to phage morphology, genomic organization, and phylogeny analysis, Vibrio phage KIT05 was classified into the family Podoviridae. The genome annotation revealed that KIT05 had no virulent or lysogenic genes. This study may help identify a novel candidate for developing biocontrol agents for Vibrio parahaemolyticus.




Similar content being viewed by others
Data Availability
The complete genome sequence of Vibrio phage KIT05 were deposited in the DNA Data Bank of Japan DDBJ database under the accession number AP019417.
References
McLaughlin JB, DePaola A, Bopp CA et al (2005) Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters. N Engl J Med 353:1463–1470. https://doi.org/10.1056/NEJMoa051594
Su YC, Liu C (2007) Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol 24:549–558. https://doi.org/10.1016/j.fm.2007.01.005
Daniels NA, MacKinnon L, Bishop R et al (2000) Vibrio parahaemolyticus infections in the United States, 1973–1998. J Infect Dis 181:1661–1666. https://doi.org/10.1086/315459
Tran L, Nunan L, Redman RM et al (2013) Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis Aquat Organ 105:45–55. https://doi.org/10.3354/dao02621
Zhang L, Orth K (2013) Virulence determinants for Vibrio parahaemolyticus infection. Curr Opin Microbiol 16:70–77. https://doi.org/10.1016/j.mib.2013.02.002
Rojas MVR, Matté MH, Dropa M et al (2011) Characterization of Vibrio parahaemolyticus isolated from oysters and mussels in São Paulo, Brazil. Rev Inst Med Trop Sao Paulo 53:201–205. https://doi.org/10.1590/S0036-46652011000400005
Stalin N, Srinivasan P (2016) Characterization of Vibrio parahaemolyticus and its specific phage from shrimp pond in Palk Strait, South East coast of India. Biologicals 44:526–533. https://doi.org/10.1016/j.biologicals.2016.08.003
Zhang H, Yang Z, Zhou Y et al (2018) Application of a phage in decontaminating Vibrio parahaemolyticus in oysters. Int J Food Microbiol 275:24–31. https://doi.org/10.1016/j.ijfoodmicro.2018.03.027
Jun JW, Han JE, Giri SS et al (2018) Phage application for the protection from acute hepatopancreatic necrosis disease (AHPND) in Penaeus vannamei. Indian J Microbiol 58:114–117. https://doi.org/10.1007/s12088-017-0694-9
Van Twest R, Kropinski AM (2009) Bacteriophage Enrichment from Water and Soil BT - Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions. In: Clokie MRJ, Kropinski AM (eds) Humana Press. Totowa, NJ, pp 15–21
Hyman P, Abedon ST (2009) Practical Methods for Determining Phage Growth Parameters BT - Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions. In: Clokie MRJ, Kropinski AM (eds) Humana Press. Totowa, NJ, pp 175–202
Stroud RM, Serwer P, Ross MJ (1981) Assembly of bacteriophage T7. dimensions of the bacteriophage and its capsids. Biophys J 36:743–757. https://doi.org/10.1016/S0006-3495(81)84763-7
Adriaenssens EM, Sullivan MB, Knezevic P et al (2020) Taxonomy of prokaryotic viruses: 2018–2019 update from the ICTV bacterial and archaeal viruses subcommittee. Arch Virol 165:1253–1260. https://doi.org/10.1007/s00705-020-04577-8
Kalatzis PG, Bastías R, Kokkari C, Katharios P (2016) Isolation and characterization of two lytic bacteriophages, φst2 and φgrn1; phage therapy application for biological control of Vibrio alginolyticus in aquaculture live feeds. PLoS ONE 11:1–18. https://doi.org/10.1371/journal.pone.0151101
Liu L, Li Y, Li S et al (2014) Comparison of next-generation sequencing systems. In: Kumar S (ed) The Role of Bioinformatics in Agriculture. Apple Academic Press, London, pp 1–25
Zerbino DR (2010) Using the Velvet de novo assembler for short-read sequencing technologies. Curr Protoc Bioinforma. https://doi.org/10.1002/0471250953.bi1105s31 (Chapter 11: Unit-11. 5)
Besemer J (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618. https://doi.org/10.1093/nar/29.12.2607
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. https://doi.org/10.1093/nar/25.5.0955
Lowe TM, Chan PP (2016) tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44:W54–W57. https://doi.org/10.1093/nar/gkw413
Garneau JR, Depardieu F, Fortier LC et al (2017) PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci Rep 7:1–10. https://doi.org/10.1038/s41598-017-07910-5
Darling ACE (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. https://doi.org/10.1101/gr.2289704
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. https://doi.org/10.1093/nar/22.22.4673
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Zuckerkandl E, Pauling L (1965) Evolutionary divergence and convergence in proteins. Evolving Genes and Proteins. Paper presented at A Symposium Held at the Institute of Microbiology of Rutgers: the State University with Support from the National Science Foundation, Academic Press, London, pp 97–166. https://www.sciencedirect.com/science/article/pii/B9781483227344500176
Letchumanan V, Chan K-G, Pusparajah P et al (2016) Insights into bacteriophage application in controlling Vibrio species. Front Microbiol. https://doi.org/10.3389/fmicb.2016.01114
Lin H, Wang J, Jiang X et al (2018) Complete genome of a novel lytic Vibrio parahaemolyticus phage VPp1 and characterization of its endolysin for antibacterial activities. J Food Prot 81:1117–1125. https://doi.org/10.4315/0362-028x.jfp-17-278
Li F, Xing S, Fu K et al (2019) Genomic and biological characterization of the Vibrio alginolyticus-infecting “Podoviridae” bacteriophage, vB_ValP_IME271. Virus Genes 55:218–226. https://doi.org/10.1007/s11262-018-1622-8
Yang M, Liang Y, Su R et al (2019) Genome characterization of the novel lytic Vibrio parahaemolyticus phage vB_VpP_BA6. Arch Virol 164:2627–2630. https://doi.org/10.1007/s00705-019-04351-5
Li F, Tian F, Li J et al (2021) Isolation and characterization of a podovirus infecting the opportunist pathogen Vibrio alginolyticus and Vibrio parahaemolyticus. Virus Res 302:198481. https://doi.org/10.1016/j.virusres.2021.198481
Prevelige PE, Cortines JR (2018) Phage assembly and the special role of the portal protein. Curr Opin Virol 31:66–73. https://doi.org/10.1016/j.coviro.2018.09.004
Marchler-Bauer A, Lu S, Anderson JB et al (2011) CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 39:D225–D229. https://doi.org/10.1093/nar/gkq1189
Marchler-Bauer A, Bo Y, Han L et al (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. https://doi.org/10.1093/nar/gkw1129
Casjens SR (2005) Comparative genomics and evolution of the tailed-bacteriophages. Curr Opin Microbiol 8:451–458. https://doi.org/10.1016/j.mib.2005.06.014
Mirzaei MK, Eriksson H, Kasuga K et al (2014) Genomic, proteomic, morphological, and phylogenetic analyses of vB-EcoP-SU10, a podoviridae phage with C3 morphology. PLoS ONE 9:1–19. https://doi.org/10.1371/journal.pone.0116294
Lin YR, Chiu CW, Chang FY, Lin CS (2012) Characterization of a new phage, termed ϕA318, which is specific for Vibrio alginolyticus. Arch Virol 157:917–926. https://doi.org/10.1007/s00705-012-1244-8
Wong H, Wang T-Y, Yang C-W et al (2019) Characterization of a lytic vibriophage VP06 of Vibrio parahaemolyticus. Res Microbiol 170:13–23. https://doi.org/10.1016/j.resmic.2018.07.003
Casjens S (2003) Prophages and bacterial genomics: what have we learned so far? Mol Microbiol 49:277–300. https://doi.org/10.1046/j.1365-2958.2003.03580.x
Yang M, Chen H, Guo S et al (2022) Characterization and genome analysis of a novel Vibrio parahaemolyticus phage vB_VpP_DE17. Virus Res 307:198580. https://doi.org/10.1016/j.virusres.2021.198580
Schmelcher M, Donovan DM, Loessner MJ (2012) Bacteriophage endolysins as novel antimicrobials. Future Microbiol 7:1147–1171. https://doi.org/10.2217/fmb.12.97
Wang W, Li M, Lin H et al (2016) The Vibrio parahaemolyticus-infecting bacteriophage qdvp001: genome sequence and endolysin with a modular structure. Arch Virol 161:2645–2652. https://doi.org/10.1007/s00705-016-2957-x
Casjens SR, Gilcrease EB (2009) Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol Biol 502:91–111. https://doi.org/10.1007/978-1-60327-565-1_7
Chung Y-B, Hinkle DC (1990) Bacteriophage T7 DNA Packaging: I. Plasmids containing a T7 replication origin and the T7 concatemer junction are packaged into transducing particles during phage infection. J Mol Biol 216:911–926. https://doi.org/10.1016/S0022-2836(99)80010-2
Krüger DH, Schroeder C (1981) Bacteriophage T3 and bacteriophage T7 virus-host cell interactions. Microbiol Rev 45:9–51. https://doi.org/10.1128/mr.45.1.9-51.1981
Nobrega FL, Vlot M, de Jonge PA et al (2018) Targeting mechanisms of tailed bacteriophages. Nat Rev Microbiol 16:760–773. https://doi.org/10.1038/s41579-018-0070-8
Cuervo A, Pulido-Cid M, Chagoyen M et al (2013) Structural characterization of the bacteriophage T7 tail machinery. J Biol Chem 288:26290–26299. https://doi.org/10.1074/jbc.M113.491209
Acknowledgements
Not applicable
Funding
This research is partially funded by the Grants-in-Aid from the Japan Society for the Promotion of Science (JSPS) Core-to-Core Program, B. Asia-Africa Science platforms.
Author information
Authors and Affiliations
Contributions
VTTA, P-KNH and KK conceived, designed and coordinated the study. VTTA and P-KNH carried out the experiment. VTTA took the lead in writing, NSH edited the English grammar of the manuscript. All authors provided critical feedback and helped shape the research, analysis and manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical Approval
This article does not describe any studies with human or other animal participants.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Anh, V.T.T., Pham-Khanh, N.H., Han, N.S. et al. Characterization and Complete Genomic Analysis of Vibrio Parahaemolyticus-Infecting Phage KIT05. Curr Microbiol 79, 221 (2022). https://doi.org/10.1007/s00284-022-02907-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-02907-4