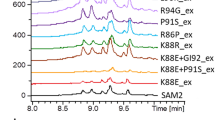
Abstract
Bacterial resistance to streptomycin is often acquired as a consequence of mutations in rpsL, the gene encoding ribosomal protein S12. Corynebacterium glutamicum is a non-pathogenic Gram-positive soil bacterium that has been widely used in industry. In a previous study, we screened several streptomycin-resistant rpsL K43 mutants of C. glutamicum, and surprisingly found that two of them also confer chloramphenicol and/or kanamycin resistance. In order to understand whether or not a single mutation of rpsLK43 could confer resistance to multiple antibiotics, in this study we attempted to construct saturation mutagenesis of rpsL K43 by rational genetic manipulation. Despite many efforts had been made, only nine mutants were successfully constructed. They were indeed resistant to streptomycin, but not to other antibiotics. This suggested that other mutations should be acquired, contributing to multiple antibiotics in the screened strains. The growth and enhanced green fluorescent protein (eGFP) expression of these nine mutants were then investigated. The results showed that they grew differently in CGXII minimal medium, but not in BHI medium. When cultured in the absence of streptomycin, the expression of eGFP was positively proportional to the growth, approximately, while in the presence of streptomycin, the expression of eGFP was proportional to the ability of streptomycin resistance.




Similar content being viewed by others
References
Davies J, Davies D (2016) Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74(3):417–433
Wang JY, Nielsen J, Liu ZH (2021) Synthetic biology advanced natural product discovery. Metabolites 11(11):785
Lewis K (2012) Antibiotics: recover the lost art of drug discovery. Nature 485(7399):439–440
Pelchovich G, Schreiber R, Zhuravlev A, Gophna U (2013) The contribution of common rpsL mutations in Escherichia coli to sensitivity to ribosome targeting antibiotics. Int J Med Microbiol 303:558–562
Islam MM, Tan Y, Hameed HMA, Chhotaray C, Liu Z, Liu Y, Lu Z, Wang S, Cai X, Gao Y, Cai X, Guo L, Li X, Tan S, Yew WW, Zhong N, Liu J, Zhang T (2020) Phenotypic and genotypic characterization of streptomycin-resistant multidrug-resistant Mycobacterium tuberculosis clinical isolates in Southern China. Microb Drug Resist 26(7):766–775
Nie L, Zhang R, Zhang L, Ma M, Li C, Zhang Y, An Y, Xu H, Xiao S, Wang T (2021) Mutations in the regulatory regions result in increased streptomycin resistance and keratinase synthesis in Bacillus thuringiensis. Arch Microbiol 203(9):5387–5396
Stella RG, Gertzen CGW, Smits SHJ, Gätgens C, Polen T, Noack S, Frunzke J (2021) Biosensor-based growth-coupling and spatial separation as an evolution strategy to improve small molecule production of Corynebacterium glutamicum. Metab Eng 68:162–173
Wendisch VF (2020) Metabolic engineering advances and prospects for amino acid production. Met Eng 58:17–34
Wang T, Li YJ, Li J, Zhang DZ, Cai NY, Zhao GH, Ma HK, Shang C, Ma Q, Xu QY, Chen N (2019) An update of the suicide plasmid-mediated genome editing system in Corynebacterium glutamicum. Microb Biotechnol 12(5):907–919
Wang Y, Hu L, Huang H, Wang H, Zhang T, Chen J, Du G, Kang Z (2020) Eliminating the capsule-like layer to promote glucose uptake for hyaluronan production by engineered Corynebacterium glutamicum. Nat Commun 11(1):3120
Eggeling L, Bott M (2005) Handbook of Corynebacterium glutamicum. CRC Press Taylor & Francis Group, Boca Raton
Wu F, Chen W, Peng Y, Tu R, Lin Y, Xing J, Wang Q (2020) Design and reconstruction of regulatory parts for fast-frowing Vibrio natriegens. ACS Synth Biol 9(9):2399–2409
Xu X, Li X, Liu Y, Zhu Y, Li J, Du G, Chen J, Ledesma-Amaro R, Liu L (2020) Pyruvate-responsive genetic circuits for dynamic control of central metabolism. Nat Chem Biol 16(11):1261–1268
Sudhir K, Glen S, Koichiro T (2016) MEGA 7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Demirci H, Murphy F, Murphy E (2013) A structural basis forstreptomycin-induced misreading of the genetic code. Nat Commun 4:1355
Wohlgemuth I, Garofalo R, Samatova E, Günenç AN, Lenz C, Urlaub H, Rodnina MV (2021) Translation error clusters induced by aminoglycoside antibiotics. Nat Commun 12(1):1830
Belardinelli R, Sharma H, Peske F, Rodnina MV (2021) Perturbation of ribosomal subunit dynamics by inhibitors of tRNA translocation. RNA 27(9):981–990
Lin JZ, Zhou DJ, Steitz TA, Polikanov YS, Gagnon MG (2018) Ribosome-targeting antibiotics: modes of action, mechanisms of resistance, and implications for drug design. Annu Rev Biochem 87(1):451–478
Loveland AB, Demo G, Grigorieff N, Korostelev AA (2017) Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature 546(7656):113–117
Loveland AB, Demo G, Korostelev AA (2020) Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading. Nature 584(7822):640–645
Hoffer ED, Hong S, Sunita S, Maehigashi T, Gonzalez RL Jr, Whitford PC, Dunham CM (2020) Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon-anticodon pairing. Elife 9:e51898
Qian H, Li J, Pan X, Sun Z, Ye C, Jin G, Fu Z (2012) Effects of streptomycin on growth of algae Chlorella vulgaris and Microcystis aeruginosa. Environ Toxicol 27(4):229–237
Rodríguez-Beltrán J, León-Sampedro R, Ramiro-Martínez P, de la Vega C, Baquero F, Levin BR, San Millán Á (2021) Translational demand is not a major source of plasmid-associated fitness costs. Philos Trans R Soc Lond, B, Biol Sci 377(1842):20200463
Gygli SM, Borrell S, Trauner A, Gagneux S (2017) Antimicrobial resistance in Mycobacterium tuberculosis: mechanistic and evolutionary perspectives. FEMS Microbiol Rev 41(3):354–373
Wu C, Paciorek M, Liu K, LeClere S, Perez-Jones A, Westra P, Sammons RD (2021) Investigating the presence of compensatory evolution in dicamba resistant IAA16 mutated kochia (Bassia scoparia). Pest Manag Sci 77(4):1775–1785
Maisnier-Patin S, Berg OG, Liljas L, Andersson DI (2002) Compensatory adaptation to the deleterious effect of antibiotic resistance in Salmonella typhimurium. Mol Microbiol 46:355–366
Maisnier-Patin S, Paulander W, Pennhag A, Andersson DI (2007) Compensatory evolution reveals functional interactions between ribosomal proteins S12, L14 and L19. J Mol Biol 366(1):207–215
Paulander W, Maisnier-Patin S, Andersson DI (2009) The fitness cost of streptomycin resistance depends on rpsL mutation, carbon source and RpoS (σS). Genetics 183(2):539–546
Gagneux S, Long CD, Small PM, Van T, Schoolnik GK, Bohannan BJM (2006) The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 312:1944–1946
O’Neill AJ, Huovinen T, Fishwick CWG, Chopra I (2006) Molecular genetic and structural modeling studies of Staphylococcus aureus RNA polymerase and the fitness of rifampin resistance genotypes in relation to clinical prevalence. Antimicrob Agents Chemother 50:298–309
Baker S, Duy PT, Nga TVT, Dung TTN, Phat VV, Chau TT, Turner AK, Farrar J, Boni MF (2013) Fitness benefits in fluoroquinolone-resistant Salmonella Typhi in the absence of antimicrobial pressure. Elife 2:e01229
Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, Galagan J, Niemann S, Gagneux S (2011) Whole-genome sequencing of rifampicin resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet 44:106–110
Levin BR, Perrot V, Walker N (2000) Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics 154(3):985–997
Björkman J, Nagaev I, Berg OG, Hughes D, Andersson DI (2000) Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 287(5457):1479–1482
Acknowledgements
This work was financially supported by the National Key Research and Development Program of China (2021YFC2100900).
Author information
Authors and Affiliations
Contributions
GHZ, RS, MY, HKM, TDB performed the study; HJ and CW analyzed the data and drafted the manuscript; YJL initiated the work, revised, and edited the manuscript. All authors approved the final version and endorsed the guidance.
Corresponding author
Ethics declarations
Conflict of Interest
All authors declare that they have no conflicts of interest.
Consent of Participants
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, G., Su, R., Yuan, M. et al. Physiological Responses of Ribosomal Protein S12 K43 Mutants of Corynebacterium glutamicum. Curr Microbiol 79, 94 (2022). https://doi.org/10.1007/s00284-022-02795-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-02795-8