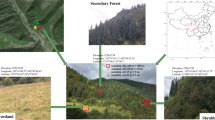
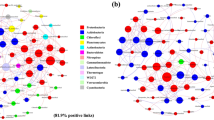
Abstract
Bacterial communities have been identified as functional key members in soil ecology. A deep relation with these communities maintains forest coverture. Trees harbor particular bacteriomes in the rhizosphere, endosphere, or phyllosphere, different from bulk-soil representatives. Moreover, the plant microbiome appears to be specific for the plant-hosting species, varies through season, and responsive to several environmental factors. This work reports the changes in bacterial communities associated with dominant pioneer trees [Tabebuia rosea and Handroanthus chrysanthus [(Bignoniaceae)] during tropical forest recovery chronosequence in the Mayan forest in Campeche, Mexico. Massive 16S sequencing approach leads to identifying phylotypes associated with rhizosphere, bulk-soil, or recovery stage. Lotka–Volterra interactome modeling suggests the presence of putative regulatory roles of some phylotypes over the rest of the community. Our results may indicate that bacterial communities associated with pioneer trees may establish more complex regulatory networks than those found in bulk-soil. Moreover, modeled regulatory networks predicted from rhizosphere samples resulted in a higher number of nodes and interactions than those found in the analysis of bulk-soil samples.




Similar content being viewed by others
Data Availability
The crude sequence dataset is available through NCBI accession number PRJNA673517. The pipeline for the bioinformatics data process is included in Supplementary File SF1. The samples analyzed may be available upon request after a share transfer agreement. The datasets generated during the current study are available from the corresponding author upon reasonable request.
References
García-Licona JB, Esparza-Olguín LG, Martínez-Romero E (2014) Estructura y composición de la vegetación leñosa de selvas en diferentes estadios sucesionales en el ejido El Carmen II, Calakmul, México. Polibotánica 38:1–26
Petersen IAB, Meyer KM, Bohannan BJM (2019) Meta-analysis reveals consistent bacterial responses to land use change across the tropics. Front Ecol Evol. https://doi.org/10.3389/fevo.2019.00391
Cobo-Díaz JF, Fernández-González AJ, Villadas PJ et al (2015) Metagenomic assessment of the potential microbial nitrogen pathways in the rhizosphere of a Mediterranean forest after a wildfire. Microb Ecol 69:895–904. https://doi.org/10.1007/s00248-015-0586-7
Mirza BS, Potisap C, Nüsslein K et al (2014) Response of free-living nitrogen-fixing microorganisms to land use change in the Amazon rainforest. Appl Environ Microbiol 80:281–288. https://doi.org/10.1128/AEM.02362-13
Ezeokoli OT, Bezuidenhout CC, Maboeta MS et al (2020) Structural and functional differentiation of bacterial communities in post-coal mining reclamation soils of South Africa: bioindicators of soil ecosystem restoration. Sci Rep 10:1759. https://doi.org/10.1038/s41598-020-58576-5
Aryal DR, De Jong BHJ, Ochoa-Gaona S et al (2014) Carbon stocks and changes in tropical secondary forests of southern Mexico. Agric Ecosyst Environ 195:220–230. https://doi.org/10.1016/j.agee.2014.06.005
Aryal DR, De Jong BHJ, Ochoa-Gaona S et al (2015) Successional and seasonal variation in litterfall and associated nutrient transfer in semi-evergreen tropical forests of SE Mexico. Nutr Cycl Agroecosyst 103:45–60. https://doi.org/10.1007/s10705-015-9719-0
van der Heijden MGA, Bardgett RD, van Straalen NM (2008) The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett 11:296–310. https://doi.org/10.1111/j.1461-0248.2007.01139.x
Madsen EL (2011) Microorganisms and their roles in fundamental biogeochemical cycles. Curr Opin Biotechnol 22:456–464. https://doi.org/10.1016/j.copbio.2011.01.008
Wood JL, Green PT, Vido JJ et al (2020) Correction to: microbial communities associated with distance- and density-dependent seedling mortality in a tropical rainforest. Plant Ecol. https://doi.org/10.1007/s11258-020-01018-z
Mishra S, Hättenschwiler S, Yang X (2020) The plant microbiome: a missing link for the understanding of community dynamics and multifunctionality in forest ecosystems. Appl Soil Ecol 145:103345. https://doi.org/10.1016/j.apsoil.2019.08.007
Fierer N, Nemergut D, Knight R, Craine JM (2010) Changes through time: integrating microorganisms into the study of succession. Res Microbiol 161:635–642. https://doi.org/10.1016/j.resmic.2010.06.002
Dukunde A, Schneider D, Schmidt M et al (2019) Tree species shape soil bacterial community structure and function in temperate deciduous forests. Front Microbiol. https://doi.org/10.3389/fmicb.2019.01519
de Habiyaremye JD, Goldmann K, Reitz T et al (2020) Tree root zone microbiome: exploring the magnitude of environmental conditions and host tree impact. Front Microbiol. https://doi.org/10.3389/fmicb.2020.00749
Shade A, Jacques M-A, Barret M (2017) Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr Opin Microbiol 37:15–22. https://doi.org/10.1016/j.mib.2017.03.010
Urbanová M, Šnajdr J, Baldrian P (2015) Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biol Biochem 84:53–64. https://doi.org/10.1016/j.soilbio.2015.02.011
de Carvalho AL, D’Oliveira MVN, Putz FE, de Oliveira LC (2017) Natural regeneration of trees in selectively logged forest in western Amazonia. For Ecol Manag 392:36–44. https://doi.org/10.1016/j.foreco.2017.02.049
Silva JO, Espírito-Santo MM, Melo GA (2012) Herbivory on Handroanthus ochraceus (Bignoniaceae) along a successional gradient in a tropical dry forest. Arthropod Plant Interact 6:45–57. https://doi.org/10.1007/s11829-011-9160-5
Dos Santos FS, De Paula RC, Sabonaro DZ, Valadares J (2009) Biometria e qualidade fisiológica de sementes de diferentes matrizes de Tabebuia chrysotricha (Mart. Ex A. DC.) Stand I. Sci For Sci 37:163–173
Pignataro AG, Levy-Tacher SI, Aguirre-Rivera JR et al (2017) Natural regeneration of tree species in pastures on peasant land in Chiapas, Mexico. Agric Ecosyst Environ 249:137–143. https://doi.org/10.1016/j.agee.2017.08.020
Horstman E, Ayón J, Griscom H (2018) Growth, survival, carbon rates for some dry tropical forest trees used in enrichment planting in the Cerro Blanco protected forest on the Ecuadorian coast. J Sustain For 37:82–96. https://doi.org/10.1080/10549811.2017.1387153
Fernandez-Vega J, Covey KR, Ashton MS (2017) Large-scale infrequent disturbances and their role in regenerating shade-intolerant tree species in Mesoamerican rainforests: implications for sustainable forest management. For Ecol Manage 395:48–68. https://doi.org/10.1016/j.foreco.2017.03.025
Gentry AH (1970) A revision of Tabebuia (Bignoniaceae) in Central America. Brittonia 22:246. https://doi.org/10.2307/2805907
Chiquini-Heredia W, Esparza-Olguín L, Peña-Ramírez Y et al (2017) Estructura y diversidad en selva inundable al centro y sur de Calakmul. Ecosist Recur Agropec 4:511–524
Chan Dzul AM (2010) Diversidad florística y funcional a través de una cronosecuencia de la selva mediana subperennifolia en la zona de influencia de la Reserva de la Biosfera Calakmul, Campeche, México. Centro Agronómico Tropical de Investigación y Enseñanza (CATIE) Turrialba, Costa Rica
Haas-Ek MA, González-Valdivia NA, De Jong BH et al (2019) Rebrote arbóreo en la regeneración del bosque tropical de Calakmul, Campeche, México. Rev Biol Trop. https://doi.org/10.15517/rbt.v67i1.33092
Gómez OC, Luiz JHH (2018) Endophytic fungi isolated from medicinal plants: future prospects of bioactive natural products from Tabebuia/Handroanthus endophytes. Appl Microbiol Biotechnol 102:9105–9119. https://doi.org/10.1007/s00253-018-9344-3
Gamboa MA, Laureano S, Bayman P (2003) Measuring diversity of endophytic fungi in leaf fragments: does size matter? Mycopathologia 156:41–45
Yarte ME, Gismondi MI, Llorente BE, Larraburu EE (2020) Isolation of endophytic bacteria from the medicinal, forestal and ornamental tree Handroanthus impetiginosus. Environ Technol. https://doi.org/10.1080/09593330.2020.1818833
Dohlman AB, Shen X (2019) Mapping the microbial interactome: statistical and experimental approaches for microbiome network inference. Exp Biol Med 244:445–458. https://doi.org/10.1177/1535370219836771
Estrada-Medina H, Canto-Canché BB, de los Santos-Briones C, O’Connor-Sánchez A (2016) Yucatán in black and red: linking edaphic analysis and pyrosequencing-based assessment of bacterial and fungal community structures in the two main kinds of soil of Yucatán State. Microbiol Res 188–189:23–33. https://doi.org/10.1016/j.micres.2016.04.007
Marquez-Santacruz HA, Hernandez-Leon R, Orozco-Mosqueda MC et al (2010) Diversity of bacterial endophytes in roots of Mexican husk tomato plants (Physalis ixocarpa) and their detection in the rhizosphere. Genet Mol Res 9:2372–2380. https://doi.org/10.4238/vol9-4gmr921
Caparso JG, Kuczyndki J, Knight R (2010) QIIME allows analysis og high-throughput community sequencing data. Nat Methods 7:335–336
Bokulich NA, Kaehler BD, Rideout JR et al (2018) Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. https://doi.org/10.1186/s40168-018-0470-z
Shaw GT-W, Pao Y-Y, Wang D (2016) MetaMIS: a metagenomic microbial interaction simulator based on microbial community profiles. BMC Bioinform 17:488. https://doi.org/10.1186/s12859-016-1359-0
Palma-López D, Zavala-Cruz J, Bautista-Zúñiga F et al (2017) Clasificación y cartografía de suelos del estado de Campeche, México. Agroproductividad 10:71–78
Pajares S, Campo J, Bohannan BJM, Etchevers JD (2018) Environmental controls on soil microbial communities in a seasonally dry tropical forest. Appl Environ Microbiol. https://doi.org/10.1128/AEM.00342-18
Rousk J, Bååth E, Brookes PC et al (2010) Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4:1340–1351. https://doi.org/10.1038/ismej.2010.58
Girvan MS, Bullimore J, Pretty JN et al (2003) Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl Environ Microbiol 69:1800–1809. https://doi.org/10.1128/AEM.69.3.1800
Ritter CD, Zizka A, Barnes C et al (2019) Locality or habitat? Exploring predictors of biodiversity in Amazonia. Ecography 42:321–333. https://doi.org/10.1111/ecog.03833
Kaiser K, Wemheuer B, Korolkow V et al (2016) Driving forces of soil bacterial community structure, diversity, and function in temperate grasslands and forests. Sci Rep 6:33696. https://doi.org/10.1038/srep33696
Bossio DA, Girvan MS, Verchot L et al (2005) Soil microbial community response to land use change in an agricultural landscape of western Kenya. Microb Ecol 49:50–62. https://doi.org/10.1007/s00248-003-0209-6
Gaby JC, Buckley DH (2011) A global census of nitrogenase diversity. Environ Microbiol 13:1790–1799. https://doi.org/10.1111/j.1462-2920.2011.02488.x
Isobe K, Koba K, Otsuka S, Senoo K (2011) Nitrification and nitrifying microbial communities in forest soils. J For Res 16:351–362. https://doi.org/10.1007/s10310-011-0266-5
Szukics U, Hackl E, Zechmeister-Boltenstern S, Sessitsch A (2012) Rapid and dissimilar response of ammonia oxidizing archaea and bacteria to nitrogen and water amendment in two temperate forest soils. Microbiol Res 167:103–109. https://doi.org/10.1016/j.micres.2011.04.002
Nelson MB, Martiny AC, Martiny JBH (2016) Global biogeography of microbial nitrogen-cycling traits in soil. Proc Natl Acad Sci USA 113:8033–8040. https://doi.org/10.1073/pnas.1601070113
Uroz S, Calvaruso C, Turpault M-P, Frey-Klett P (2009) Mineral weathering by bacteria: ecology, actors and mechanisms. Trends Microbiol 17:378–387. https://doi.org/10.1016/j.tim.2009.05.004
Bergkemper F, Schöler A, Engel M et al (2016) Phosphorus depletion in forest soils shapes bacterial communities towards phosphorus recycling systems. Environ Microbiol 18:1988–2000. https://doi.org/10.1111/1462-2920.13188
Hu X, Chen J, Guo J (2006) Two phosphate- and potassium-solubilizing bacteria isolated from Tianmu Mountain, Zhejiang, China. World J Microbiol Biotechnol 22:983–990. https://doi.org/10.1007/s11274-006-9144-2
Brown ME, Chang MC (2014) Exploring bacterial lignin degradation. Curr Opin Chem Biol 19:1–7. https://doi.org/10.1016/j.cbpa.2013.11.015
Esson KC, Lin X, Kumaresan D et al (2016) Alpha- and gammaproteobacterial methanotrophs codominate the active methane-oxidizing communities in an acidic boreal peat bog. Appl Environ Microbiol 82:2363–2371. https://doi.org/10.1128/AEM.03640-15
Uroz S, Calvaruso C, Turpault MP et al (2007) Effect of the mycorrhizosphere on the genotypic and metabolic diversity of the bacterial communities involved in mineral weathering in a forest soil. Appl Environ Microbiol 73:3019–3027. https://doi.org/10.1128/AEM.00121-07
Izumi H, Cairney JWG, Killham K et al (2008) Bacteria associated with ectomycorrhizas of slash pine (Pinus elliottii) in south-eastern Queensland, Australia. FEMS Microbiol Lett 282:196–204. https://doi.org/10.1111/j.1574-6968.2008.01122.x
Yu H-X, Wang C-Y, Tang M (2013) Fungal and bacterial communities in the rhizosphere of Pinus tabulaeformis related to the restoration of plantations and natural secondary forests in the Loess Plateau, Northwest China. Sci World J 2013:1–12. https://doi.org/10.1155/2013/606480
Kai M, Effmert U, Berg G, Piechulla B (2007) Volatiles of bacterial antagonists inhibit mycelial growth of the plant pathogen Rhizoctonia solani. Arch Microbiol 187:351–360. https://doi.org/10.1007/s00203-006-0199-0
Kai M, Haustein M, Molina F et al (2009) Bacterial volatiles and their action potential. Appl Microbiol Biotechnol 81:1001–1012. https://doi.org/10.1007/s00253-008-1760-3
Lladó S, López-Mondéjar R, Baldrian P (2017) Forest soil bacteria: diversity, involvement in ecosystem processes, and response to global change. Microbiol Mol Biol Rev. https://doi.org/10.1128/MMBR.00063-16
Kawasaki A, Watson ER, Kertesz MA (2012) Indirect effects of polycyclic aromatic hydrocarbon contamination on microbial communities in legume and grass rhizospheres. Plant Soil 358:169–182. https://doi.org/10.1007/s11104-011-1089-z
Štursová M, Žifčáková L, Leigh MB et al (2012) Cellulose utilization in forest litter and soil: identification of bacterial and fungal decomposers. FEMS Microbiol Ecol 80:735–746. https://doi.org/10.1111/j.1574-6941.2012.01343.x
Větrovský T, Steffen KT, Baldrian P (2014) Potential of cometabolic transformation of polysaccharides and lignin in lignocellulose by soil actinobacteria. PLoS ONE 9:e89108. https://doi.org/10.1371/journal.pone.0089108
Rösch C, Mergel A, Bothe H (2002) Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl Environ Microbiol 68:3818–3829. https://doi.org/10.1128/AEM.68.8.3818-3829.2002
Jamalizadeh M, Etebarian HR, Aminian H, Alizadeh A (2010) Biological control of Botrytis mali on apple fruit by use of Bacillus bacteria, isolated from the rhizosphere of wheat. Arch Phytopathol Plant Prot 43:1836–1845. https://doi.org/10.1080/03235400902830960
López-Mondéjar R, Zühlke D, Becher D et al (2016) Cellulose and hemicellulose decomposition by forest soil bacteria proceeds by the action of structurally variable enzymatic systems. Sci Rep 6:25279. https://doi.org/10.1038/srep25279
Nacke H, Thürmer A, Wollherr A et al (2011) Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS ONE. https://doi.org/10.1371/journal.pone.0017000
Knelman JE, Legg TM, O’Neill SP et al (2012) Bacterial community structure and function change in association with colonizer plants during early primary succession in a glacier forefield. Soil Biol Biochem 46:172–180. https://doi.org/10.1016/j.soilbio.2011.12.001
Redford AJ, Bowers RM, Knight R et al (2010) The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ Microbiol 12:2885–2893. https://doi.org/10.1111/j.1462-2920.2010.02258.x
Haichar FEZ, Marol C, Berge O et al (2008) Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2:1221–1230. https://doi.org/10.1038/ismej.2008.80
Wertz S, Degrange V, Prosser JI et al (2007) Decline of soil microbial diversity does not influence the resistance and resilience of key soil microbial functional groups following a model disturbance. Environ Microbiol 9:2211–2219. https://doi.org/10.1111/j.1462-2920.2007.01335.x
Griffiths B, Ritz K, Wheatley R et al (2001) An examination of the biodiversity–ecosystem function relationship in arable soil microbial communities. Soil Biol Biochem 33:1713–1722. https://doi.org/10.1016/S0038-0717(01)00094-3
Bardgett R, Bowman W, Kaufmann R, Schmidt S (2005) A temporal approach to linking aboveground and belowground ecology. Trends Ecol Evol 20:634–641. https://doi.org/10.1016/j.tree.2005.08.005
Garcia J, Kao-Kniffin J (2019) Can dynamic network modelling be used to identify adaptive microbiomes? Funct Ecol. https://doi.org/10.1111/1365-2435.13491
Pascale A, Proietti S, Pantelides IS, Stringlis IA (2020) Modulation of the root microbiome by plant molecules: the basis for targeted disease suppression and plant growth promotion. Front Plant Sci. https://doi.org/10.3389/fpls.2019.01741
Trivedi P, Leach JE, Tringe SG et al (2020) Plant–microbiome interactions: from community assembly to plant health. Nat Rev Microbiol 18:607–621. https://doi.org/10.1038/s41579-020-0412-1
Pedrinho A, Mendes LW, Merloti LF et al (2019) Forest-to-pasture conversion and recovery based on assessment of microbial communities in Eastern Amazon rainforest. FEMS Microbiol Ecol. https://doi.org/10.1093/femsec/fiy236
Porter-Bolland L, Ellis EA, Gholz HL (2007) Land use dynamics and landscape history in La Montaña, Campeche, Mexico. Landsc Urban Plan 82:198–207. https://doi.org/10.1016/j.landurbplan.2007.02.008
López-Lozano NE, Echeverría Molinar A, Ortiz Durán EA et al (2020) Bacterial diversity and interaction networks of agave lechuguilla rhizosphere differ significantly from bulk soil in the oligotrophic basin of Cuatro Cienegas. Front Plant Sci. https://doi.org/10.3389/fpls.2020.01028
Acknowledgements
This work was funded by El Colegio de la Frontera Sur (Project #5103711808) and by Consejo Estatal de Investigación Científica y Desarrollo Tecnológico del Estado de Campeche (COESICYDET). AABL thanks Consejo Nacional de Ciencia y Tecnología (CONACYT) for its Master’s scholarship number 624546.
Funding
Consejo Estatal de Investigación Científica y Desarrollo Tecnológico de Campeche (COESICYDET), Research Grant YPR 2015-2018. El Colegio de la Frontera Sur, Research Grant 5103711808. AABL Consejo Nacional de Ciencia y Tecnología, Scholarship Grant 624546.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation and data collection and analysis were performed by all authors. The first draft of the manuscript was written by Yuri Jorge Peña-Ramírez, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
This work received approval from the “Comité de Ética para la Investigación” from El Colegio de la Frontera Sur in April 2017. Samples were collected after approval from SEMARNAT according to “Permiso de colecta de material biológico para investigación científica” SGPA/DGGFS/712/2062/17 granted to the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Becerra-Lucio, A.A., Labrín-Sotomayor, N.Y., Becerra-Lucio, P.A. et al. Diversity and Interactomics of Bacterial Communities Associated with Dominant Trees During Tropical Forest Recovery. Curr Microbiol 78, 3417–3429 (2021). https://doi.org/10.1007/s00284-021-02603-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-021-02603-9