Abstract
Xanthomonas oryzae pv. oryzae (X. oryzae) is a bacterial pathovar of rice diseases all over the world. Owing to emerging antibacterial resistance, phage therapies have gained significant attention to treat various bacterial infections. Nevertheless, comprehensive research is needed for their use as a safe biocontrol agent. In this study, isolation and characterization of a novel phage Xoo-sp15, that infects X. oryzae was ascertained through experimental and bioinformatics analyses to determine its virulent potency and reliability. High throughput sequencing demonstrated that Xoo-sp15 has a dsDNA genome with a total size of 157,091 bp and 39.9% GC content lower than its host (63.6%). Morphological and phylogenetic analyses characterized it as a new member of the Bastille-like group within the family Herelleviridae. In silico analysis revealed that it contains 229 open reading frames and 16 tRNAs. Additionally, this novel phage does not contain any resistant determinants and can infect nine X. oryzae strains. Therefore, Xoo-sp15 has the potential to serve as a novel candidate for phage therapy.



Similar content being viewed by others
References
Liu W, Liu J, Triplett L, Leach JE, Wang G-L (2014) Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu Rev Phytopathol 52:213–241
Lee B-M, Park Y-J, Park D-S, Kang H-W, Kim J-G, Song E-S, Park I-C, Yoon U-H, Hahn J-H, Koo B-S (2005) The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res 33(2):577–586
Fahad S, Nie L, Khan FA, Chen Y, Hussain S, Wu C, Xiong D, Jing W, Saud S, Khan FA (2014) Disease resistance in rice and the role of molecular breeding in protecting rice crops against diseases. Biotech Lett 36(7):1407–1420
Dong Z, Xing S, Liu J, Tang X, Ruan L, Sun M, Tong Y, Peng D (2018) Isolation and characterization of a novel phage Xoo-sp2 that infects Xanthomonas oryzae pv. oryzae. J Gen Virol 99(10):1453–1462
Young R, Gill JJ (2015) Phage therapy redux—What is to be done? Science 350(6265):1163–1164
Xi H, Dai J, Tong Y, Cheng M, Zhao F, Fan H, Li X, Cai R, Ji Y, Sun C (2019) The characteristics and genome analysis of vB_AviM_AVP, the first phage infecting aerococcus viridans. Viruses 11(2):104
Jurczak-Kurek A, Gąsior T, Nejman-Faleńczyk B, Bloch S, Dydecka A, Topka G, Necel A, Jakubowska-Deredas M, Narajczyk M, Richert M (2016) Biodiversity of bacteriophages: morphological and biological properties of a large group of phages isolated fr om urban sewage. Sci Rep. https://doi.org/10.1038/srep34338
Inoue Y, Matsuura T, Ohara T, Azegami K (2006) Bacteriophage OP 1, lytic for Xanthomonas oryzae pv. oryzae, changes its host range by duplication and deletion of the small domain in the deduced tail fiber gene. J Gen Plant Pathol 72(2):111–118
Inoue Y, Matsuura T, Ohara T, Azegami K (2006) Sequence analysis of the genome of OP 2, a lytic bacteriophage of Xanthomonas oryzae pv. oryzae. J Gen Plant Pathol 72(2):104–110
Salzberg SL, Sommer DD, Schatz MC, Phillippy AM, Rabinowicz PD, Tsuge S, Furutani A, Ochiai H, Delcher AL, Kelley D (2008) Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99 A. BMC Genomics 9(1):204
Yang B, Bogdanove A (2013) Inoculation and virulence assay for bacterial blight and bacterial leaf streak of rice. Rice Protocols. Springer, New York, pp 249–255
Van Twest R, Kropinski AM (2009) Bacteriophage enrichment from water and soil. Bacteriophages. Springer, New York, pp 15–21
MrA S (1991) An improved method for the small scale preparation of bacteriophage DNA based on phage precipitation by zinc chloride. Nucleic Acids Res 19(19):5442
Xie Y, Wu G, Tang J, Luo R, Patterson J, Liu S, Huang W, He G, Gu S, Li S (2014) SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 30(12):1660–1666
Li S, Fan H, An X, Fan H, Jiang H, Chen Y, Tong Y (2014) Scrutinizing virus genome termini by high-throughput sequencing. PLoS ONE 9(1):e85806
Stephen FA (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25(5):955–964
Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV (2012) Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67(11):2640–2644
Sullivan MJ, Petty NK, Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinformatics 27(7):1009–1010
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Barylski J, Enault F, Dutilh BE, Schuller MB, Edwards RA, Gillis A, Klumpp J, Knezevic P, Krupovic M, Kuhn JH (2020) Analysis of spounaviruses as a case study for the overdue reclassification of tailed phages. Syst Biol 69(1):110–123
Ogunyemi SO, Chen J, Zhang M, Wang L, Masum MMI, Yan C, An Q, Li B, Chen J (2019) Identification and characterization of five new OP2-related Myoviridae bacteriophages infecting different strains of Xanthomonas oryzae pv. oryzae. J Plant Pathol 101(2):263–273
El-Arabi TF, Griffiths MW, She Y-M, Villegas A, Lingohr EJ, Kropinski AM (2013) Genome sequence and analysis of a broad-host range lytic bacteriophage that infects the Bacillus cereus group. Virol J 10(1):48
Hatfull GF, Hendrix RW (2011) Bacteriophages and their genomes. Curr Opin Virol 1(4):298–303
Guilliam TA, Keen BA, Brissett NC, Doherty AJ (2015) Primase-polymerases are a functionally diverse superfamily of replication and repair enzymes. Nucleic Acids Res 43(14):6651–6664
Greenstein D, Zinder ND, Horiuchi K (1988) Integration host factor interacts with the DNA replication enhancer of filamentous phage f1. Proc Natl Acad Sci 85(17):6262–6266
Flower AM, McHenry CS (1990) The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc Natl Acad Sci 87(10):3713–3717
Zhang L, Xu D, Huang Y, Zhu X, Rui M, Wan T, Zheng X, Shen Y, Chen X, Ma K (2017) Structural and functional characterization of deep-sea thermophilic bacteriophage GVE2 HNH endonuclease. Sci Rep 7:42542
O’Flaherty S, Coffey A, Edwards R, Meaney W, Fitzgerald G, Ross R (2004) Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G+ C content. J Bacteriol 186(9):2862–2871
Heichlinger A, Ammelburg M, Kleinschnitz E-M, Latus A, Maldener I, Flärdh K, Wohlleben W, Muth G (2011) The MreB-like protein Mbl of streptomyces coelicolor A3 (2) depends on MreB for proper localization and contributes to spore wall synthesis. J Bacteriol 193(7):1533–1542
Schwarzer D, Stummeyer K, Haselhorst T, Freiberger F, Rode B, Grove M, Scheper T, von Itzstein M, Mühlenhoff M, Gerardy-Schahn R (2009) Proteolytic release of the intramolecular chaperone domain confers processivity to endosialidase F. J Biol Chem 284(14):9465–9474
Adams RLP, Burdon RH (1985) The function of DNA methylation in bacteria and phage. Molecular biology of DNA methylation. Springer New York, New York, NY, pp 73–87. https://doi.org/10.1007/978-1-4612-5130-9_6
Casjens S, Hendrix R (1988) Control mechanisms in dsDNA bacteriophage assembly. The bacteriophages. Springer, Boston, pp 15–91
Fokine A, Rossmann MG (2016) Common evolutionary origin of procapsid proteases, phage tail tubes, and tubes of bacterial type VI secretion systems. Structure 24(11):1928–1935
North OI, Sakai K, Yamashita E, Nakagawa A, Iwazaki T, Büttner CR, Takeda S, Davidson AR (2019) Phage tail fibre assembly proteins employ a modular structure to drive the correct folding of diverse fibres. Nature Microbiol 4(10):1645–1653
Lucks JB, Nelson DR, Kudla GR, Plotkin JB (2008) Genome landscapes and bacteriophage codon usage. PLoS Comput Biol 4(2):e1000001
Limor-Waisberg K, Carmi A, Scherz A, Pilpel Y, Furman I (2011) Specialization versus adaptation: two strategies employed by cyanophages to enhance their translation efficiencies. Nucleic Acids Res 39(14):6016–6028
Bahir I, Fromer M, Prat Y, Linial M (2009) Viral adaptation to host: a proteome-based analysis of codon usage and amino acid preferences. Mol Syst Biol 5(1):311
Bailly-Bechet M, Vergassola M, Rocha E (2007) Causes for the intriguing presence of tRNAs in phages. Genome Res 17(10):1486–1495
Kuo T-T, Huang T-C, Chow T-Y (1969) A filamentous bacteriophage from Xanthomonas oryzae. Virology 39(3):548–555
Yuzenkova J, Nechaev S, Berlin J, Rogulja D, Kuznedelov K, Inman R, Mushegian A, Severinov K (2003) Genome of Xanthomonas oryzae bacteriophage Xp10: an odd T-odd phage. J Mol Biol 330(4):735–748
Lee C-N, Hu R-M, Chow T-Y, Lin J-W, Chen H-Y, Tseng Y-H, Weng S-F (2007) Comparison of genomes of three Xanthomonas oryzae bacteriophages. BMC Genomics 8(1):442
Ji Z, Ji C, Liu B, Zou L, Chen G, Yang B (2016) Interfering TAL effectors of Xanthomonas oryzae neutralize R-gene-mediated plant disease resistance. Nat Commun 7(1):1–9
Acknowledgements
Special thanks to the authors Zhaoxia Dong and Jin Liu for providing help in this study.
Funding
Funding was supported by YFA0903000 (Grant No. YFA0903000).
Author information
Authors and Affiliations
Contributions
D.P, H.Q., and Y.T. proposed the idea and designed the experiments. Z.D. and J.L performed the experiments. A.N performed the computational analyses, drafted and wrote the manuscript. R.A.T and N.A critically read, analyzed and revised the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
All the authors declared that there are no conflicts of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
284_2021_2556_MOESM1_ESM.tif
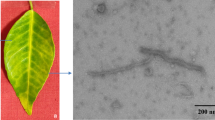
Supplementary file1 Phage morphology of Xoo-sp15. Phage was negatively stained with 2% phosphotungstic acid (PTA) and examined by transmission electron microscopy (TEM) at an accelerating voltage of 200 kV. The scale bar represents 200 nm. (TIF 9562 kb)
284_2021_2556_MOESM2_ESM.tif
Supplementary file2 Morphology of Xoo-sp15 plaques. Phages were plated in Nutrient Broth agar and overlain with a liquid culture of PXO99A. The plates were incubated at 37 °C. Clear, well-defined Xoo-sp15 plaques were observed and photographed after 36 h. (TIF 1484 kb)
284_2021_2556_MOESM3_ESM.tif
Supplementary file3 Genetic and physical organization of Xoo-sp15 genome. The 229 ORFs of Xoo-sp15 are depicted, and the direction of transcription is indicated by arrows. The G + C content and skew of Xoo-sp15 are also shown. The circle map of the Xoo-sp15 genome was made using CGView (http://wishart.biology.ualberta.ca/cgview/). (TIF 3141 kb)
284_2021_2556_MOESM4_ESM.jpg
Supplementary file4 Comparative genomic analysis of Xoo-sp15 and its homologues. Phages that share over best-hit proteins with Xoo-sp15 were selected and compared using the Easyfig tool. (JPG 3208 kb)
Rights and permissions
About this article
Cite this article
Nazir, A., Dong, Z., Liu, J. et al. Isolation, Characterization, and Genome Sequence Analysis of a Novel Lytic Phage, Xoo-sp15 Infecting Xanthomonas oryzae pv. oryzae. Curr Microbiol 78, 3192–3200 (2021). https://doi.org/10.1007/s00284-021-02556-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-021-02556-z