Abstract
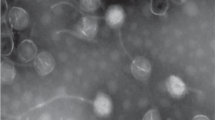
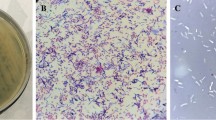
Although bacteriophages are more numerous and have smaller genomes than their bacterial hosts, relatively few have their genomes sequenced. Here, we isolated the Pseudomonas fluorescens bacteriophage from Napahai plateau wetland and performed de novo genome sequencing. Based on the previous biological characteristics and bioinformatics analysis, it was determined that VW-6B was a linear double-stranded DNA (dsDNA) phage with 35,306 bp, with 56.76% G+C content and 197 bp tandem repeats. The VW-6B genome contained 46 open-reading frames (ORFs), and no tRNA genes were found. Based on phage genome structure, sequence comparison, and collinear analysis, VW-6B should be classified into the family Siphoviridae and be considered as a member of a new species in the Mu-like phage. The newly isolated bacteriophage can specifically infect P. fluorescens, which further enriches the diversity of known bacteriophages and provides a basis for the subsequent research and application of bacteriophages.




Similar content being viewed by others
Data Availability
The datasets generated or analyzed during the current study are available from the corresponding author on reasonable request.
References
Suttle CA (2007) Marine viruses–major players in the global ecosystem. Nat Rev Microbiol 5(10):801–812. https://doi.org/10.1038/nrmicro1750
Hoai TD, Nishiki I, Fujiwara A, Yoshida T, Nakai T (2019) Comparative genomic analysis of three lytic lactococcus garvieae phages, novel phages with genome architecture linking the 936 phage species of lactococcus lactis. Mar Genomics. https://doi.org/10.1016/j.margen.2019.100696
Salisbury A, Tsourkas PK (2019) A method for improving the accuracy and efficiency of bacteriophage genome annotation. Int J Mol Sci. https://doi.org/10.3390/ijms20143391
Perez Sepulveda B, Redgwell T, Rihtman B, Pitt F, Scanlan DJ, Millard A (2016) Marine phage genomics: the tip of the iceberg. FEMS Microbiol Lett. https://doi.org/10.1093/femsle/fnw158
Hatfull GF (2015) Dark matter of the biosphere: the amazing world of bacteriophage diversity. J Virol 89(16):8107–8110. https://doi.org/10.1128/JVI.01340-15
Zhao Y, Temperton B, Thrash JC, Schwalbach MS, Vergin KL, Landry ZC, Ellisman M, Deerinck T, Sullivan MB, Giovannoni SJ (2013) Abundant SAR11 viruses in the ocean. Nature 494(7437):357–360. https://doi.org/10.1038/nature11921
Montano SP, Pigli YZ, Rice PA (2012) The mu transpososome structure sheds light on DDE recombinase evolution. Nature 491(7424):413–417. https://doi.org/10.1038/nature11602
Meyer JR, Dobias DT, Weitz JS, Barrick JE, Quick RT, Lenski RE (2012) Repeatability and contingency in the evolution of a key innovation in phage lambda. Science 335(6067):428–432. https://doi.org/10.1126/science.1214449
Bradley DE (1963) The structure of coliphages. J Gen Microbiol 31:435–445. https://doi.org/10.1099/00221287-31-3-435
Bradley DE (1966) The fluorescent staining of bacteriophage nucleic acids. J Gen Microbiol 44(3):383–391. https://doi.org/10.1099/00221287-44-3-383
Tolstoy I, Kropinski AM, Brister JR (2018) Bacteriophage taxonomy: an evolving discipline. Methods Mol Biol 1693:57–71. https://doi.org/10.1007/978-1-4939-7395-8_6
Piligrimova EG, Kazantseva OA, Nikulin NA, Shadrin AM (2019) Bacillus phage vB_BtS_B83 previously designated as a plasmid may represent a new siphoviridae genus. Viruses-Basel 11(7):624. https://doi.org/10.3390/V11070624
Faelen M, Toussaint A (1976) Bacteriophage Mu-1: a tool to transpose and to localize bacterial genes. J Mol Biol 104(3):525–539. https://doi.org/10.1016/0022-2836(76)90118-2
Wang X, Higgins NP (1994) “Muprints” of the lac operon demonstrate physiological control over the randomness of in vivo transposition. Mol Microbiol 12(4):665–677. https://doi.org/10.1111/j.1365-2958.1994.tb01054.x
Bukhari AI, Froshauer S, Botchan M (1976) Ends of bacteriophage mu DNA. Nature 264(5586):580–583. https://doi.org/10.1038/264580a0
Harshey RM (2012) The Mu story: how a maverick phage moved the field forward. Mob DNA 3(1):21. https://doi.org/10.1186/1759-8753-3-21
Hulo C, Masson P, Le Mercier P, Toussaint A (2015) A structured annotation frame for the transposable phages: a new proposed family “Saltoviridae” within the caudovirales. Virology 477:155–163. https://doi.org/10.1016/j.virol.2014.10.009
Lee SY, Dunn RJK, Young RA, Connolly RM, Dale PER, Dehayr R, Lemckert CJ, McKinnon S, Powell B, Teasdale PR, Welsh DT (2006) Impact of urbanization on coastal wetland structure and function. Austral Ecol 31(2):149–163. https://doi.org/10.1111/j.1442-9993.2006.01581.x
Qin K, Ji X, Zhang C, Ding Y, Kuang A, Zhang S, Zhang Q, Lin L, Wei Y (2017) Isolation and characterization of wetland VSW-3, a novel lytic cold-active bacteriophage of pseudomonas fluorescens. Can J Microbiol 63(2):110–118. https://doi.org/10.1139/cjm-2016-0368
Cai R, Wang Z, Wang G, Zhang H, Cheng M, Guo Z, Ji Y, Xi H, Wang X, Xue Y, Ur Rahman S, Sun C, Feng X, Lei L, Tong Y, Han W, Gu J (2019) Biological properties and genomics analysis of vB_KpnS_GH-K3, a Klebsiella phage with a putative depolymerase-like protein. Virus Genes. https://doi.org/10.1007/s11262-019-01681-z
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25(5):955–964. https://doi.org/10.1093/nar/25.5.955
Teng T, Li Q, Liu Z, Li X, Liu Z, Liu H, Liu F, Xie L, Wang H, Zhang L, Wu D, Chen M, Li Y, Ji A (2019) Characterization and genome analysis of novel klebsiella phage henu1 with lytic activity against clinical strains of klebsiella pneumoniae. Adv Virol 164(9):2389–2393. https://doi.org/10.1007/s00705-019-04321-x
Wang D, Jiang Y, Xiao S, Wang M, Liu Q, Huang L, Xue C, Wang Q, Lin T, Shao H, McMinn A (2019) Characterization and genome analysis of a novel alteromonas phage JH01 isolated from the qingdao coast of China. Curr Microbiol. https://doi.org/10.1007/s00284-019-01751-3
Braid MD, Silhavy JL, Kitts CL, Cano RJ, Howe MM (2004) Complete genomic sequence of bacteriophage B3, a Mu-like phage of pseudomonas aeruginosa. J Bacteriol 186(19):6560–6574. https://doi.org/10.1128/JB.186.19.6560-6574.2004
Akhwale JK, Rohde M, Rohde C, Bunk B, Sproer C, Klenk HP, Boga HI, Wittmann J (2019) Comparative genomic analysis of eight novel haloalkaliphilic bacteriophages from lake elmenteita. Kenya PloS one 14(2):e0212102. https://doi.org/10.1371/journal.pone.0212102
Xiang Y, Wang S, Li J, Wei Y, Zhang Q, Lin L, Ji X (2018) Isolation and characterization of two lytic cold-active bacteriophages infecting pseudomonas fluorescens from the napahai plateau wetland. Can J Microbiol 64(3):183–190. https://doi.org/10.1139/cjm-2017-0572
Koberg S, Gieschler S, Brinks E, Wenning M, Neve H, Franz C (2018) Genome sequence of the novel virulent bacteriophage PMBT14 with lytic activity against pseudomonas fluorescens DSM 50090(R). Arch Virol 163(9):2575–2577. https://doi.org/10.1007/s00705-018-3882-y
Li M, Chen X, Ma Y, Li Z, Zhao Q (2018) Complete genome sequence of PFP1, a novel T7-like pseudomonas fluorescens bacteriophage. Arch Virol 163(12):3423–3426. https://doi.org/10.1007/s00705-018-3979-3
Sillankorva S, Neubauer P, Azeredo J (2008) Isolation and characterization of a T7-like lytic phage for pseudomonas fluorescens. BMC Biotechnol 8:80. https://doi.org/10.1186/1472-6750-8-80
Zhang C, Zhang Z, Li J, Qin K, Wei Y, Zhang Q, Lin L, Ji X (2017) Complete genome sequence of the lytic cold-active pseudomonas fluorescens bacteriophage VSW-3 from napahai plateau wetland. Virus Genes 53(1):146–150. https://doi.org/10.1007/s11262-016-1403-1
Poullain V, Gandon S, Brockhurst MA, Buckling A, Hochberg ME (2008) The evolution of specificity in evolving and coevolving antagonistic interactions between a bacteria and its phage. Evolution 62(1):1–11. https://doi.org/10.1111/j.1558-5646.2007.00260.x
Eller MR, Salgado RL, Vidigal PM, Alves MP, Dias RS, de Oliveira LL, da Silva CC, de Carvalho AF, De Paula SO (2013) Complete genome sequence of the pseudomonas fluorescens bacteriophage UFV-P2. Genome Announc. https://doi.org/10.1128/genomeA.00006-12
Lu G, Luhr J, Stoecklein A, Warner P, Tapprich W (2017) Complete genome sequences of pseudomonas fluorescens bacteriophages isolated from freshwater samples in omaha nebraska. Genome. https://doi.org/10.1128/genomeA.01501-16
Casjens S (2003) Prophages and bacterial genomics: what have we learned so far? Mol Microbiol 49(2):277–300. https://doi.org/10.1046/j.1365-2958.2003.03580.x
Casjens SR, Gilcrease EB (2009) Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol Biol 502:91–111. https://doi.org/10.1007/978-1-60327-565-1_7
Abedon ST (2009) Phage evolution and ecology. Adv Appl Microbiol 67:1–45. https://doi.org/10.1016/S0065-2164(08)01001-0
Lima-Mendez G, Toussaint A, Leplae R (2011) A modular view of the bacteriophage genomic space: identification of host and lifestyle marker modules. Res Microbiol 162(8):737–746. https://doi.org/10.1016/j.resmic.2011.06.006
Berry JD, Rajaure M, Young R (2013) Spanin function requires subunit homodimerization through intermolecular disulfide bonds. Mol Microbiol 88(1):35–47. https://doi.org/10.1111/mmi.12167
Summer EJ, Berry J, Tran TA, Niu L, Struck DK, Young R (2007) Rz/Rz1 lysis gene equivalents in phages of gram-negative hosts. J Mol Biol 373(5):1098–1112. https://doi.org/10.1016/j.jmb.2007.08.045
Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Ruger W (2003) Bacteriophage T4 genome. Microbiol Mol Biol Rev. https://doi.org/10.1128/mmbr.67.1.86-156.2003
Morgan GJ, Hatfull GF, Casjens S, Hendrix RW (2002) Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in haemophilus, Neisseria and Deinococcus. J Mol Biol 317(3):337–359. https://doi.org/10.1006/jmbi.2002.5437
Roberts MD, Martin NL, Kropinski AM (2004) The genome and proteome of coliphage T1. Virology 318(1):245–266. https://doi.org/10.1016/j.virol.2003.09.020
Acknowledgements
This research was supported by the National Natural Science Foundation of China (31860147 and 31700324). XJ, YW, QZ, and KQ designed research; ZC and ZX performed research; ZC, ZX, and XJ analyzed data and wrote the paper.
Funding
The National Natural Science Foundation of China (31860147 and 31700324).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
I would like to declare on behalf of my co-authors that the work described was original research that has not been published previously, and not under consideration for publication elsewhere in whole or in part.
Consent to Participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for Publication
Manuscript is approved by all authors for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cui, Z., Xu, Z., Wei, Y. et al. Characterization and Genome Analysis of a Novel Mu-like Phage VW-6B Isolated from the Napahai Plateau Wetland of China. Curr Microbiol 78, 150–158 (2021). https://doi.org/10.1007/s00284-020-02277-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02277-9