Abstract
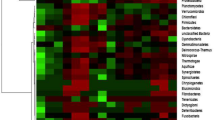
Changme Khangpu glacier is located in the northern district of Sikkim which comes under UNESCO heritage site Kanchenjunga Biosphere Reserve which is considered as one of the important biological hotspot regions in the Eastern Himalayas. This is the first report on microbial diversity analysis of moraine soil from one of the unexplored glaciers of Sikkim using high throughput sequencing platform and phospholipid fatty acids analysis (PLFA). It was found that the 16S amplicon sequence comprised 362,902 raw sequences with a sequence length of 150 bp and (G + C) content 52%. A total of 156,821 pre-processed reads were clustered into 378 OTUs (operational taxonomic units) comprising 6 bacterial phyla. The top four dominant phyla based on the 16S amplicon sequences were Proteobacteria (56%), Firmicutes (16%), Actinobacteria (12%), and Bacteroidetes (8%), respectively. PLFA analysis confirmed the dominance of Gram positive bacteria (72%) followed by Gram negative bacteria (32%) and the major fatty acids which are present in the moraine soil sample were PUFA (61%), and 18:2ω6,9c (29%). This is the primary study and first of its kind done on moraine soil from glaciers of Sikkim. Based on 16S amplicon sequencing and PLFA analysis of moraine soil samples from glaciers of Sikkim suggest that this glaciers harbours rich microbial diversity and thus can have wide industrial and biotechnological potential. Thus, there is an escalating scope to further study these extreme biomes with respect to their microbial diversity and their functional capabilities.





Similar content being viewed by others
References
Hamdan A (2018) Psychrophiles: ecological significance and potential industrial application. S Afr J Sci 114:1–6. https://doi.org/10.17159/sajs.2018/20170254
Rampelotto PH (2013) Extremophiles and extreme environments. Life 3:482–485. https://doi.org/10.3390/life3030482
Sherpa MT, Najar IN, Das S, Thakur N (2018) Bacterial diversity in an alpine debris-free and debris-cover accumulation zone glacier ice, North Sikkim, India. Indian J Microbiol 58:470–478. https://doi.org/10.1007/s12088-018-0747-8
Christner BC, Mosley-Thompson E, Thompson LG, Reeve JN (2003) Bacterial recovery from ancient glacial ice. Environ Microbiol 5:433–436. https://doi.org/10.1046/j.1462-2920.2003.00422.x
Wang Z, Luo T, Li R, Tang Y, Du M (2013) Causes for the unimodal pattern of biomass and productivity in alpine grasslands along a large altitudinal gradient in semi-arid regions. J Veg Sci 24:189–201. https://doi.org/10.1111/j.1654-1103.2012.01442.x
Priscu JC, Christner BC (2014) Earth’s icy biosphere earth’s icy biosphere 13:130–145
Priscu JC, Adams EE, Lyons WB, Voytek M, Mogk D, Brown RL, McKay CP, Takacs CD, Welch KA, Wolf C, Kirshtein JD, Avci R (1999) Geomicrobiology of subglacial ice above Lake Vostok, Antarctica. Science 286:2141–2144. https://doi.org/10.1126/s-cience.286.5447.2141
Sheridan PP, Miteva VI, Brenchley JE (2003) Phylogenetic analysis of anaerobic psychrophilic enrichment cultures obtained from a Greenland glacier ice core. Appl Environ Microbiol 69:2153–2160. https://doi.org/10.1128/AEM.69.4.2153-2160.200-3
Miteva VI, Sheridan PP, Brenchley JE (2004) Phylogenetic and physiological diversity of microorganisms isolated from a deep Greenland glacier ice core. Appl Environ Microbiol 70:202–213. https://doi.org/10.1128/AEM.70.1.202-213.2004
Xiang S, Yao T, An L, Xu B (2005) 16S rRNA sequences and differences in bacteria isolated from the Muztag Ata glacier at increasing depths 16S rRNA sequences and differences in bacteria isolated from the Muztag Ata glacier at increasing depths. Appl Environ Microbiol 71:4619–4627. https://doi.org/10.1128/AEM.71.8.4619-4627.2005
Helmke E, Weyland H (2004) Psychrophilic versus psychrotolerant bacteria—occurrence and significance in polar and temperate marine habitats. Cell Mol Biol (Noisy-le-grand) 50:553–561. https://doi.org/10.1170/T545
Fan F, Zhang B, Morrill PL (2017) Phospholipid fatty acid (PLFA) analysis for profiling microbial communities in offshore produced water. Mar Pollut Bull 122:194–206. https://doi.org/10.1016/j.marpolbul.2017.06.044
Vartoukian SR, Palmer RM, Wade WG (2010) Strategies for culture of “unculturable” bacteria. FEMS Microbiol Lett 309:1–7. https://doi.org/10.1111/j.1574-6968-2010.02-000.x
Pham VHT, Kim J (2012) Cultivation of unculturable soil bacteria. Trends Biotechnol 30:475–484. https://doi.org/10.1016/j.marpolbul.2017.06.044
Zhang X, Ma X, Yao T, Zhang G (2003) Diversity of 16S rDNA and environmental factor influncing microorganisms in Malan ice core. Chinese Sci Bull 48:1146–1151. https://doi.org/10.1007/BF03185770
Foght J, Aislabie J, Turner S, Brown CE, Ryburn J, Saul DJ, Lawson W (2004) Culturable bacteria in subglacial sediments and ice from two Southern Hemisphere glaciers. Microb Ecol 47:329–340. https://doi.org/10.1007/s00248-003-1036-5
Willers C, Jansen van Rensburg PJ, Claassens S (2015) Microbial signature lipid biomarker analysis—an approach that is still preferred, even amid various method modifications. J Appl Microbiol 118:1251–1263. https://doi.org/10.1111/jam.12789
Kellenberger E (2001) Exploring the unknown the silent revolution of microbiology. EMBO Rep 2:5–7. https://doi.org/10.1093/embo-reports/kve014
Sherpa MT, Najar IN, Das S, Thakur N (2019) Culture independent bacterial diversity of Changme Khang and Changme Khangpu glaciers of North Sikkim, India. Environ Sustain. https://doi.org/10.1007/s42398-019-00067-z
Sherpa MT, Najar IN, Das S, Thakur N (2020) Distribution of antibiotic and metal resistance genes in two glaciers of North Sikkim, India. Ecotoxicol Environ Saf 203:111037. https://doi.org/10.1016/j.ecoenv.2020.1111037
Quideau SA, McIntosh ACS, Norris CE, Lloret E, Methew JB, Swallow HK (2016) Extraction and analysis of microbial phospholipid fatty acids in soils. J Vis Exp. https://doi.org/10.3791/54360
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glockner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:1–11. https://doi.org/10.1093/nar/gks808
Faircloth BC, Glenn TC, White ND (2014) Illumina library prep protocol. CCA 4.0 United States Licens 1–15
Najar IN, Sherpa MT, Das S, Das S, Thakur N (2018) Microbial ecology of two hot springs of Sikkim: predominate population and geochemistry. Sci Total Environ 637–638:730–745. https://doi.org/10.1016/j.scitotenv.2018.05.037
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. https://doi.org/10.1038/nmeth.2604
Basnett S, Kulkarni AV, Arrawatia ML, Shrestha DG (2016) Glacier studies in Sikkim Himalaya. https://doi.org/10.13140/RG.2.2.24023.88487
Anesio AM, Hodson AJ, Fritz A, Psenner R, Sattler B (2009) High microbial activity on glaciers: importance to the global carbon cycle. Glob Chang Biol 15:955–960. https://doi.org/10.1111/j.1365-2486.2008.01758.x
Hodson A, Anesio AM, Tranter M, Fountain A, Osborn M, Prisu J, Laybourn-Parry J, Sattler B (2016) Glacial ecosystems. Ecol Monogr 78:41–67. https://doi.org/10.1890/07-0187.1
Junge K, Christner B, James TS (2011) Diversity of psychrophilic bacteria from sea ice—and glacial ice communities. Extrem Handb. https://doi.org/10.1007/9-784-431-538981-39
Jones HG (1999) The ecology of snow-covered systems: a brief overview of nutrient cycling and life in the cold. Hydrol Process 13:2135–2147. https://doi.org/10.1002/(SI-CI)10991085(199910)13:14/15%3c2135::AIDHYP862%3e3.0.CO;2-Y
Liu Y, Yao T, Jiao N, Kang S, Zeng Y, Huang S (2006) Microbial community structure in moraine lakes and glacial meltwaters, Mount Everest. FEMS Microbiol Lett 265:98–105. https://doi.org/10.1111/j.1574-6968.2006.00477.X
Larose C, Berger S, Ferrari C, Navarro E, Dommergue A, Schneider D, Vogel TM (2010) Microbial sequences retrieved from environmental samples from seasonal Arctic snow and meltwater from Svalbard, Norway. Extremophiles 14:205–212. https://doi.org/10.1007/s00792-009-0299-2
Hell K, Edwards A, Zarsky J, Podmirseg SM, Girdwood SE, Pachebat J, Insam H, Sattler B (2013) The dynamic bacterial communities of a melting high arctic glacier snowpack. ISME J 7:1814–1826. https://doi.org/10.1038/ismej.2013.51
Lopatina A, Krylenkov V, Severinov K (2013) Activity and bacterial diversity of snow around russian antarctic stations. Res Microbiol 164:949–958. https://doi.org/10.1016/j.resmic.2013.08.005
Møller AK, Søborg DA, Al-Soud WA, Sørensen SJ, Kroer N (2013) Bacterial community structure in high-arctic snow and freshwater as revealed by pyrosequencing of 16S rRNA genes and cultivation. Polar Res. https://doi.org/10.3402/polar.v32i-0.17390
Cheng SM, Foght JM (2007) Cultivation-independent and -dependent characterization of bacteria resident beneath John Evans glacier. FEMS Microbiol Ecol 59:318–330. https://doi.org/10.1111/j.1574-6941.2006.00267.x
Segawa T, Miyamoto K, Ushida K, Agata K, Okada N, Kohshima S (2005) Seasonal change in bacterial flora and biomass in mountain snow from the Tateyama Mountains, Japan, analyzed by 16S rRNA gene sequencing and real-time PCR. Appl Environ Microbiol 71:123–130. https://doi.org/10.1128/AEM.71.1.123-130.2005
Willerslev E, Hansen AJ, Poinar HN (2004) Isolation of nucleic acids and cultures from fossil ice and permafrost. Trends Ecol Evol 19:141–147. https://doi.org/10.1016/j.tre-e.2003.11.010
Turchetti B, Buzzini P, Goretti M, Branda E, Diolaiuti G, D’Agata SC, Vaughan-Martini A (2008) Psychrophilic yeasts in glacial environments of Alpine glaciers. FEMS Microbiol Ecol 63:73–83. https://doi.org/10.1111/j.15746941.2007.0-0409.x
Skidmore M, Anderson SP, Sharp M, Foght J, Lanoil BD (2005) Comparison of microbial community compositions of two subglacial environments reveals a possible role for microbes in chemical weathering processes. Appl Environ Microbiol 71:6986–6997. https://doi.org/10.1128/AEM.71.11.6986-6997.2005
Mosier AC, Murray AE, Fritsen CH (2007) Microbiota within the perennial ice cover of Lake Vida, Antarctica. FEMS Microbiol Ecol 59:274–288. https://doi.org/10.1111/j.1574-6941.2006.00220.x
Acknowledgements
The authors would like to thank the Department of Forest, Govt. of Sikkim for kind permission and Department of Microbiology, Sikkim University for laboratory space.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no competing interests.
Nucleotide sequence accession number
Metagenome sequence data are available at NCBI Accession no. SRP165759 (direct link to deposition data: https://www.ncbi.nlm.nih.gov/sra/SRX4887442).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sherpa, M.T., Najar, I., Das, S. et al. Exploration of Microbial Diversity of Himalayan Glacier Moraine Soil Using 16S Amplicon Sequencing and Phospholipid Fatty Acid Analysis Approaches. Curr Microbiol 78, 78–85 (2021). https://doi.org/10.1007/s00284-020-02259-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02259-x