Abstract
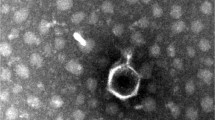
A novel virulent phage, vB_KpnP_IME337, isolated from a hospital sewage in Beijing, China, that infects carbapenem-resistant Klebsiella pneumoniae KN2 capsular type was identified and characterized. Next-generation sequencing and genome analysis revealed that vB_KpnP_IME337 had a linear double-stranded genome with a length of 44,266 base pairs and G+C content of 53.7%. Fifty-two putative open reading frames were identified, and no transfer RNA-encoding genes were detected. BLASTn analysis revealed that phage vB_KpnP_IME337 had the highest sequence similarity with Klebsiella phage phiBO1E, with genome coverage of 79%. Based on morphology, phage vB_KpnP_IME337 was determined to belong to the family Podoviridae of the order Caudovirales. It was shown that phage vB_KpnP_IME337 had an infection duration of ~ 90 min and 10 min latent period, and a highly specific to host strain. In conclusion, phage vB_KpnP_IME337 may be a promising alternative candidate to antibiotic treatment for controlling diseases caused by drug-resistant K. pneumoniae.



Similar content being viewed by others
References
Renshaw A (2009) Pulmonary pathology, a volume in the foundations in diagnostic pathology series. Adv Anat Pathol 16(1):65
Podschun R, Ullmann U (1998) Klebsiella spp as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11(4):589–603
Cunningham AA, Daszak P, Wood JLN (2017) One Health, emerging infectious diseases and wildlife: two decades of progress? Philos Trans R Soc Lond Ser B 372(1725):20160167
Zilberberg MD, Shorr AF, Micek ST, Vazquez-Guillamet C, Kollef MHJCC (2014) Multi-drug resistance, inappropriate initial antibiotic therapy and mortality in Gram-negative severe sepsis and septic shock: a retrospective cohort study. Crit Care 18(6):1–13
Souli M, Galani I, Antoniadou A, Papadomichelakis E, Poulakou G, Panagea T, Vourli S, Zerva L, Armaganidis A, Kanellakopoulou K, Giamarellou H (2010) An outbreak of infection due to beta-Lactamase Klebsiella pneumoniae Carbapenemase 2-producing K pneumoniae in a Greek University Hospital: molecular characterization, epidemiology, and outcomes. Clin Infect Dis 50(3):364–373
Stapleton PJ, Murphy M, Mccallion N, Brennan M (2016) Outbreaks of extended spectrum beta-lactamaseproducing Enterobacteriaceae in neonatal intensive care units: a systematic review. Arch Dis Childhood Fetal Neonatal Ed 101(1):403–403
Wisgrill L, Lepuschitz S, Blaschitz M, Rittenschober-Böhm J, Diab-El Schahawi M, Schubert S, Indra A, Berger AJPIDJ (2018) Outbreak of yersiniabactin-producing Klebsiella pneumoniae in a neonatal intensive care unit. Pediatr Infect Dis J 38(6):638–642
Ventola CLJP (2015) The antibiotic resistance crisis: part 1: causes and threats. Pharm Ther 40(4):277–283
Payne RJH, Jansen VAAJCP (2000) Phage therapy: the peculiar kinetics of self-replicating pharmaceuticals. Pharmacol Ther 68(3):225–230
Nilsson AS (2014) Phage therapy—constraints and possibilities. Upsala J Med Sci 119(2):192–198
Gill JJ, Hollyer T, Sabour PM (2007) Bacteriophages and phage-derived products as antibacterial therapeutics. Expert Opin Ther Patents 17(11):1341–1350
Matsuzaki S, Rashel M, Uchiyama J, Sakurai S, Ujihara T, Kuroda M, Ikeuchi M, Tani T, Fujieda M (2005) Bacteriophage therapy: a revitalized therapy against bacterial infectious diseases. J Infect Chemother 11(5):211–219
Domingo-Calap P, Delgado-Martinez J (2018) Bacteriophages: protagonists of a post-antibiotic era. Antibiotics (Basel, Switzerland) 7(3):66
Cha K, Oh HK, Jang JY, Jo Y, Kim WK, Ha GU, Ko KS, Myung H (2018) Characterization of two novel bacteriophages infecting multidrug-resistant (MDR) Acinetobacter baumannii and evaluation of their therapeutic efficacy in vivo. Front Microbiol 9:696
Sao-Jose C (2018) Engineering of phage-derived lytic enzymes: improving their potential as antimicrobials. Antibiotics (Basel, Switzerland) 7(2):29
Pallavali RR, Degati VL, Lomada D, Reddy MC, Durbaka VRP (2017) Isolation and in vitro evaluation of bacteriophages against MDR-bacterial isolates from septic wound infections. PLoS ONE 12(7):e0179245
Chadha P, Katare OP, Chhibber S (2017) Liposome loaded phage cocktail: enhanced therapeutic potential in resolving Klebsiella pneumoniae mediated burn wound infections. Burns 43(7):1532–1543
Manohar P, Tamhankar AJ, Lundborg CS, Nachimuthu R (2019) Therapeutic characterization and efficacy of bacteriophage cocktails infecting Escherichia coli, Klebsiella pneumoniae, and enterobacter species. Front Microbiol 10:574
Greisen K, Loeffelholz M, Purohit A, Leong D (1994) PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol 32(2):335–351
Pan YJ, Lin TL, Chen YH, Hsu CR, Hsieh PF, Wu MC, Wang JT (2013) Capsular types of Klebsiella pneumoniae revisited by wzc sequencing. PLoS ONE 8(12):e80670
Zhao F, Sun H, Zhou X, Liu G, Li M, Wang C, Liu S, Zhuang Y, Tong Y, Ren H (2019) Characterization and genome analysis of a novel bacteriophage vB_SpuP_Spp16 that infects Salmonella enterica serovar pullorum. Virus Genes 55(4):532–540
Anderson TF (1949) Bacteriophages. 4(4):21
Ackermann HW (2009) Basic phage electron microscopy. Methods Mol Biol (Clifton, NJ) 501:113–126
Delbrück MJ (1940) The growth of bacteriophage and lysis of the host. J Gen Physiol 23(5):643
Adams MH (1959) Bacteriophage. Interscience Publishers, New York, pp 450–456
Li F, Xing S, Fu K, Zhao S, Liu J, Tong Y, Zhou L (2019) Genomic and biological characterization of the Vibrio alginolyticus-infecting "Podoviridae" bacteriophage, vB_ValP_IME271. Virus Genes 55(2):218–226
Chen Y, Sun E, Song J, Yang L, Wu B (2018) Complete genome sequence of a novel T7-like bacteriophage from a pasteurella multocida capsular type A isolate. Curr Microbiol 75(5):574–579
Yang Y, Cai L, Ma R, Xu Y, Tong Y, Huang Y, Jiao N, Zhang R (2017) A novel roseosiphophage isolated from the oligotrophic South China Sea. Viruses 9(5):109
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437(7057):376–380
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75
Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305(3):567–580
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25(5):955–964
Sullivan MJ, Petty NK, Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinformatics (Oxford, England) 27(7):1009–1010
Underwood AP, Mulder A, Gharbia S, Green J (2005) Virulence searcher: a tool for searching raw genome sequences from bacterial genomes for putative virulence factors. Clin Microbiol Infect 11(9):770–772
Kleinheinz KA, Joensen KG, Larsen MV (2014) Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage 4(1):e27943
Adriaenssens EM, Wittmann J, Kuhn JH, Turner D, Sullivan MB, Dutilh BE, Jang HB, van Zyl LJ, Klumpp J, Lobocka M, Moreno Switt AI, Rumnieks J, Edwards RA, Uchiyama J, Alfenas-Zerbini P, Petty NK, Kropinski AM, Barylski J, Gillis A, Clokie MRC, Prangishvili D, Lavigne R, Aziz RK, Duffy S, Krupovic M, Poranen MM, Knezevic P, Enault F, Tong Y, Oksanen HM, Rodney Brister J (2018) Taxonomy of prokaryotic viruses: 2017 update from the ICTV bacterial and archaeal viruses subcommittee. Adv Virol 163(4):1125–1129
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 60. Mol Biol Evol 30(12):2725–2729
Zhang X, Wang Y, Li S, An X, Pei G, Huang Y, Fan H, Mi Z, Zhang Z, Wang W, Chen Y, Tong Y (2015) A novel termini analysis theory using HTS data alone for the identification of Enterococcus phage EF4-like genome termini. BMC Genomics 16:414
Mewes HW, Frishman D, Mayer KF, Munsterkotter M, Noubibou O, Pagel P, Rattei T, Oesterheld M, Ruepp A, Stumpflen V (2006) MIPS: analysis and annotation of proteins from whole genomes in 2005. Nucleic Acids Res 34:D169–D172
Shi Y, Yan Y, Ji W, Du B, Meng X, Wang H, Sun JJVJ (2012) Characterization and determination of holin protein of Streptococcus suis bacteriophage SMP in heterologous host. Virol J 9(1):70–70
Sun S, Rao VB, Biol MGRJCOS (2010) Genome packaging in viruses. Curr Opin Struct Biol 20(1):114–120
Guilliam TA, Keen BA, Brissett NC, Doherty AJJ (2015) Primase-polymerases are a functionally diverse superfamily of replication and repair enzymes. Nucleic Acids Res 43(14):6651–6664
Tu J, Park T, Morado DR, Hughes KT, Molineux IJ, Liu JJV (2017) Dual host specificity of phage SP6 is facilitated by tailspike rotation. Virology 507:206–215
Hambly E, Tetart F, Desplats C, Wilson WH, Krisch HM (2001) A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proc Natl Acad Sci 98(20):11411–11416
Acknowledgements
The work was supported by the Capital Characteristic Clinic Project of Beijing (Z161100000516181) and the National Key Research and Development Program of China (2016YFC1202705, AWS16J020, and AWS15J006) and the National Science and Technology Major Project (2018ZX10201001).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gao, M., Wang, C., Qiang, X. et al. Isolation and Characterization of a Novel Bacteriophage Infecting Carbapenem-Resistant Klebsiella pneumoniae. Curr Microbiol 77, 722–729 (2020). https://doi.org/10.1007/s00284-019-01849-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-019-01849-8