Abstract
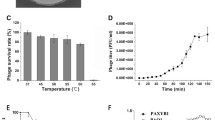
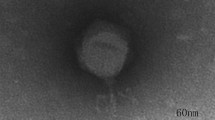
Phage PA-YS35 is a novel lytic Pseudomonas aeruginosa phage belonging to the Myoviridae family and was isolated from the sewage of the First Hospital of Jilin University. The biological properties testing indicated that phage PA-YS35 is stable between − 20 and 60 °C and pH 4–9. The one-step growth curve shows that the latent period of PA-YS35 was 9 min, and the burst period was about 21 min by the size of approximately 380 progeny phages per host cell. The genome of phage PA-YS35 is linear double-stranded DNA with a size of 93,296 bp and a GC content of 49.35%. The results from RAST gene annotation analysis showed that the PA-YS35 genome contains 172 open reading frames (ORFs); the function of 41 ORFs can be predicted, whereas the product of remaining 131 ORFs are hypothetical proteins. According to phylogenetic tree of RNA ligase encoding sequence, phage PA-YS35 has a close evolutionary relationship with Pseudomonas phage PAK P1 because both of them are located on the same branch. The study of phage PA-YS35 genome will provide useful information for further research on the interaction between phages and their hosts.






Similar content being viewed by others
References
Gould IM, Wise R (1985) Pseudomonas aeruginosa: clinical manifestations and management. The Lancet 2(8466):1224–1227
Lyczak JB et al (2000) Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect 2(9):1051–1060
Currie AJ et al (2003) Pseudomonas aeruginosa: role in the pathogenesis of the CF lung lesion. Semin Respir Crit Care Med 24:671–680
Kosorok MR et al (2001) Acceleration of lung disease in children with cystic fibrosis after Pseudomonas aeruginosa acquisition. Pediatr Pulmonol 32:277–287
Balter M (2000) Virology. Evolution on life's fringes. Science 289(5486):1866–1867
Yan L et al (2016) Complete genomic sequence of bacteriophage H188: a novel Vibrio kanaloae, phage isolated from Yellow Sea. Curr Microbiol 72(5):1–6
Sabouri Ghannad M, Mohammadi A (2012) Bacteriophage: time to re-evaluate the potential of phage therapy as a promising agent to control multidrug-resistant bacteria. Iran Basic Med Sci 15(2):693–701
Lin L et al (2010) Isolation and characterization of an extremely long tail Thermus bacteriophage from Tengchong hot springs in China. J Basic Microbiol 50(5):452–456
Middelboe M et al (2010) Isolation and lifecycle characterization of lytic viruses infecting heterotrophicbacteria and cyanobacteria. Manual of aquatic viral ecology. ASLO, Waco, pp 149–180
Duhaime MB et al (2011) Ecogenomics and genome landscapes of marine Pseudoalteromonas phage H105/1. ISME J 5(1):107–121
Hyman P, Abedon ST (2010) Bacteriophage host range andbacterial resistance. Adv Appl Microbiol 70:217–248
Li Y et al (2016) Complete genomic sequence of bacteriophage H188: a novel Vibrio kanaloae phage isolated from yellow sea. Curr Microbiol 72(5):628–633
Capra ML et al (2004) Thermal and chemical resistance of Lactobacillus casei, and Lactobacillus paracasei bacteriophages. Lett Appl Microbiol 38(6):499–504
Pajunen M et al (2010) BacteriophageuYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J Bacteriol 182(18):5114–5120
Cuervo A et al (2013) Structural characterization of the bacteriophage T7 tail machinery. J Biol Chem 288(36):26290–26299
Filée J et al (2005) Marine T4-type bacteriophages, a ubiquitous component of the dark matter of the biosphere. Proc Natl Acad Sci USA 102(35):12471–12476
Skurnik M, Strauch E (2006) Phage therapy: facts and fiction. Int J Med Microbiol 296(1):5–14
Twort FW (1936) Further investigations on the nature of ultra-microscopic viruses and their cultivation. J Hyg 36(2):204–235
Kutter E et al (2010) Phage therapy in clinical practice: treatment of human infections. Curr Pharm Biotechnol 11(1):69–86
Marchler-Bauer A et al (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45(D1):D200–D203
Marchler-Bauer A et al (2015) CDD: NCBI's conserved domain database. Nucleic Acids Res 43:222–226
Marchler-Bauer A et al (2011) CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 39(D):225–229
Marchler-Bauer A, Bryant SH (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32:327–331
Acknowledgements
This work was in part supported by the Bethune Researching Plan of Jilin university (2018B38), the Natural Science Foundation of Science and Technology Department of Jilin Province (20190201032JC), the Fundamental Research Funds for the Central Universities. Project of health Department Key Laboratory, Jilin province (2018J062).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical Approval
Although YS 35 is a clinical isolated P aeruginosa, the phage isolation and identification against it is a pure scientific process without any relationship with the patient's privacy. And there are no agents that were used in any patients. So there are no ethical problems in the research.
Research Involving Human or Animal Participants
This article does not contain any research involving human or animal participants.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jiang, YH., Liu, JQ., Zhao, CY. et al. Isolation and Genome Sequencing of a Novel Pseudomonas aeruginosa Phage PA-YS35. Curr Microbiol 77, 123–128 (2020). https://doi.org/10.1007/s00284-019-01792-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-019-01792-8