Abstract
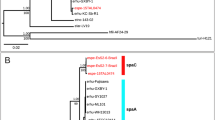
We report here the novel species to encompass the isolate A649T (=CBAS 716T = CBRVS P1061T) obtained from viscera of the healthy pufferfish Sphoeroides spengleri (Family Tetraodontidae). Genomic taxonomy analysis demonstrates that the novel strain A649T had < 95% average amino acid identity/average nucleotide identity (AAI/ANI) and < 70% similarity of genome-to-genome distance (GGDH) towards its closest neighbors which places A649T into a new Enterovibrio species (Enterovibrio baiacu sp nov.). In silico phenotyping disclosed several features that may be used to differentiate related Enterovibrio species. The nearly complete genome assembly of strain A649T consisted of 5.4 Mbp and 4826 coding genes.


Similar content being viewed by others
Data Availability
Enterovibrio baiacu A649T is deposited in the Bacteria Collection of Environmental and Health (CBAS) at the Oswaldo Cruz Institute (IOC), Oswaldo Cruz Foundation (FIOCRUZ) in Rio de Janeiro, Brazil (https://cbas.fiocruz.br/), under the accession number CBAS 716T, and in the Reference Bacterial Collection for Sanitary Surveillance (CBRVS), INCQS, under the accession number CBRVS P1061T. The whole-genome shotgun project for Enterovibrio baiacu A649T (= CBAS 716 T = CBRVS P1061T) have been deposited in GenBank under accession number SCHN00000000. The BioProject accession number is PRJNA515211.
References
Thompson F, Gomez-Gil B (2017) International Committee on Systematics of Prokaryotes Subcommittee on the taxonomy of Aeromonadaceae. Vibrionaceae and related organisms Minutes of the meeting vol. 68. Chicago, IJSEM, pp 2111–2112
Thompson FL, Iida T, Swings J (2004) Biodiversity of vibrios. Microbiol Mol Biol Rev 68:403–431
Thompson FL, Hoste B, Thompson CC, Goris J, Gomez-Gil B, Huys L, De Vos P, Swings J (2002) Enterovibrio norvegicus gen. nov., sp. nov., isolated from the gut of turbot (Scophthalmus maximus) larvae: a new member of the family Vibrionaceae. Int J Syst Evol Microbiol 52(6):2015–2022
Sawabe T, Kita-Tsukamoto K, Thompson FL (2007) Inferring the evolutionary history of vibrios by means of multilocus sequence analysis. J Bacteriol 189:7932–7936
Walter JM, Tschoeke DA, Meirelles PM, de Oliveira L, Leomil L, Tenório M, Valle R, Salomon PS, Thompson CC, Thompson FL (2016) Taxonomic and functional metagenomic signature of turfs in the abrolhos reef system (Brazil). PLoS ONE 11:e0161168
Schmieder R, Edwards R (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864
Coil D, Jospin G, Darling AE (2015) A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31:587–589
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Gen Res 9:868–877
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T et al (2014) The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:206–214
Konstantinidis KT, Tiedje JM (2005) Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci 102:2567–2572
MeierKolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60
Thompson CC, Chimetto L, Edwards RA, Swings J, Stackebrandt E, Thompson FL (2013) Microbial genomic taxonomy. BMC Genomics 14:913
De Vos P, Thompson F, Thompson C, Swings J (2017) A flavor of prokaryotic taxonomy: systematics revisited. In: Microbial resources, pp. 29–44
Amaral GRS, Dias GM, WellingtonOguri M, Chimetto L, Campeão ME, Thompson FL, Thompson CC (2014) Genotype to phenotype: identification of diagnostic vibrio phenotypes using whole genome sequences. IJSEM 64:357–365
Acknowledgements
The authors thank CNPQ, CAPES, FAPERJ, and FAPESP (Grants Number 2013/50228-8 and 2016/14375-4) for financial support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Azevedo, G.P.R., Mattsson, H.K., Appolinario, L.R. et al. Enterovibrio baiacu sp. nov.. Curr Microbiol 77, 154–157 (2020). https://doi.org/10.1007/s00284-019-01785-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-019-01785-7