Abstract
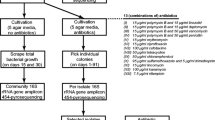
Despite the broad assessment of sponge bacterial diversity through cultivation-independent and dependent strategies, the knowledge focusing on cultivable anaerobes from this holobiont is still incipient. Plakina is a genus with the highest number of described species from the smallest of poriferan classes, Homoscleromorpha. The Brazilian Atlantic coast has been presenting itself as a hotspot for the discovery of new plakinidae species, with initial surveys just now concerning to characterize their microbiome. The current study aimed to isolate and identify strict anaerobes from recently described species of Plakina collected at the coast of Cabo Frio, RJ. Samples of four sympatric morphotypes of Plakina cyanorosea and Plakina cabofriense were collected on the coast of Cabo Frio, RJ. Using five different culture media, a total of 93 bacterial isolates were recovered, among which 60 were strict anaerobes and, ultimately, 34 remaining viable. A total of 76.5% from these strains were mostly identified as Clostridium bifermentans by mass spectrometry and 82.4% identified by 16S rRNA sequencing, almost all of them affiliated to the genus Paraclostridium, and with one isolate identified as Clostridium butyricum by both techniques. None of the anaerobic bacteria exhibited antimicrobial activity by the adopted screening test. The present work highlights not only the need for cultivation and characterization of the anaerobic microbiota from marine sponges but also adds the existing scarce knowledge of culturable bacterial communities from Homoscleromorph sponges from Brazilian coast.


Similar content being viewed by others
References
Hatti-Kaul R, Mattiasson B (2016) Anaerobes in industrial- and environmental biotechnology. In: Hatti-Kaul R, Mamo G, Mattiasson B (eds) Anaerobes in biotechnology Advances in biochemical engineering/biotechnology. Springer, Cham, pp 1–33. https://doi.org/10.1007/10_2016_10
Lloyd-Price J, Abu-Ali G, Huttenhower C (2016) The healthy human microbiome. Genom Med 8:51. https://doi.org/10.1186/s13073-016-0307-y
Haas F, Konig H (1987) Characterisation of an anaerobic symbiont and the associated aerobic bacterial flora of Pyrrhocoris apterus (Heteroptera: Pyrrhocoridae). FEMS Microbiol Ecol 45(2):99–106. https://doi.org/10.1016/0378-1097(87)90031-0
Thong-On A, Suzuki K, Noda S, Inoue J, Kajiwara S, Ohkuma M (2012) Isolation and characterization of anaerobic bacteria for symbiotic recycling of uric acid nitrogen in the gut of various termites. Microbes Environ 27(2):186–192. https://doi.org/10.1264/jsme2.ME11325
Babarro J, De Zwaan A (2008) Anaerobic survival potential of four bivalves from different habitats. A comparative survey. Comp Biochem Physiol A 151(1):108–113. https://doi.org/10.1016/j.cbpa.2008.06.006
Brück W, Brück T, Self W, Reed J, Nitecki S, McCarthy P (2010) Comparison of the anaerobic microbiota of deep-water Geodia spp. and sandy sediments in the Straits of Florida. ISME J 4(5):686–699. https://doi.org/10.1038/ismej.2009.149
Thomas T, Moitinho-Silva L, Lurgi M, Bjo¨rk JR, Easson C, Astudillo-Garcı´a C, Olson JB, Erwin PM, Lopez-Legentil S, Luter H, Chaves-Fonnegra A, Costa R, Schupp PJ, Steindler L, Erpenbeck D, Gilbert J, Knight R, Ackermann G, Victor-Lopez J, Taylor MW, Thacker RW, Montoya JM, Hentschel U, Webster NS (2016) Diversity, structure and convergent evolution of the global sponge microbiome. Nat Commun 16:11870. https://doi.org/10.1038/ncomms11870
Hentschel U, Piel J, Degnan S, Taylor M (2012) Genomic insights into the marine sponge microbiome. Nat Rev Microbiol 10(9):641–654. https://doi.org/10.1038/nrmicro2839
Hoffmann F, Larsen O, Thiel V, Rapp HT, Pape T, Michaelis W, Reitner J (2005) An anaerobic world of sponges. Geomicrobiol J 22(1–2):1–10. https://doi.org/10.1080/01490450590922505
Hoffmann F, Røy H, Bayer K, Hentschel U, Pfannkuchen M, Breummer F, Beer D (2008) Oxygen dynamics and transport in the Mediterranean sponge Aplysina aerophoba. Mar Biol 153(6):1257–1264. https://doi.org/10.1007/s00227-008-0905-3
Schumann-Kindel G, Bergbauer M, Manz W, Szewyk U, Reitner J (1997) Aerobic and anaerobic microorganisms in modern sponges: a possible relationship to fossilization-processes. Facies 36:268–272
Moitinho-Silva L, Díez-Vives C, Batani G, Esteves A, Jahn M, Thomas T (2017) Integrated metabolism in sponge–microbe symbiosis revealed by genome-centered metatranscriptomics. ISME J 11(7):1651–1666. https://doi.org/10.1038/ismej.2017.25
Pita L, Rix L, Slaby B, Franke A, Hentschel U (2018) The sponge holobiont in a changing ocean: from microbes to ecosystems. Microbiome 6(46):1. https://doi.org/10.1186/s40168-018-0428-1
Laport M (2018) Isolating bacteria from sponges: why and how? Curr Pharm Biotechnol 18(15):1224–1236. https://doi.org/10.2174/1389201019666180329111327
Lavy A, Keren R, Haber M, Schwartz I, Ilan M (2013) Implementing sponge physiological and genomic information to enhance the diversity of its culturable associated bacteria. FEMS Microbiol Ecol 87(2):486–502. https://doi.org/10.1111/1574-6941.12240
Boury-Esnault N, Lavrov DV, Ruiz CA, Pérez T (2013) The integrative taxonomic approach applied to Porifera: a case study of the Homoscleromorpha. Integr Comp Biol 53(3):416–427. https://doi.org/10.1093/icb/ict042
Domingos C, Lage A, Muricy G (2016) Overview of the biodiversity and distribution of the Class Homoscleromorpha in the Tropical Western Atlantic. J Mar Biol Assoc UK 96(2):379–389. https://doi.org/10.1017/S0025315415000375
Domingos C, Moraes F, Muricy G (2013) Four new species of Plakinidae (Porifera: Homoscleromorpha) from Brazil. Zootaxa 3718(6):530–544. https://doi.org/10.11646/zootaxa.3718.6.2
Muricy G, Domingos C, Lage A, Lanna E, Hardoim C, Laport M, Zilberberg C (2018) Integrative taxonomy widens our knowledge of the diversity, distribution and biology of the genus Plakina (Homosclerophorida: Plakinidae). Invertebr Syst. https://doi.org/10.1071/IS18027
Laport M, Bauwens M, de Oliveira Nunes S, Willenz P, George I, Muricy G (2017) Culturable bacterial communities associated to Brazilian Oscarella species (Porifera: Homoscleromorpha) and their antagonistic interactions. Antonie Van Leeuwenhoek 110(4):489–499. https://doi.org/10.1007/s10482-016-0818-y
Rodrigues N, Bronzato G, Santiago G, Botelho L, Moreira B, Coelho I, Souza M, Coelho S (2017) The Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) identification versus biochemical tests: a study with enterobacteria from a dairy cattle environment. Braz J Microbiol 48(1):132–138. https://doi.org/10.1016/j.bjm.2016.07.025
Laport MS, Santos-Gandelman JF, Muricy G Giambiagi-deMarval M, George I (2016) Antagonistic interactions among bacteria isolated from either the same or from different sponges native to the Brazilian coast. J Mar Sci Res Dev 6:185. https://doi.org/10.4172/2155-9910.1000185
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173(2):697–703. https://doi.org/10.1128/jb.173.2.697-703.1991
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35(21):7188–7196. https://doi.org/10.1093/nar/gkm864
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msy096
Marinho PR, Moreira AP, Pellegrino FL, Muricy G, Bastos MC, Santos KR, Giambiagi-deMarval M, Laport MS (2009) Marine Pseudomonas putida: a potential source of antimicrobial substances against antibiotic-resistant bacteria. MIOC 104(5):678–682. https://doi.org/10.1590/S0074-02762009000500002
Kim M, Oh HS, Park SC, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64(Pt 2):346–351. https://doi.org/10.1099/ijs.0.059774-0
Imhoff JF, Truper HG (1976) Marine sponges as habitats of anaerobic phototrophic bacteria. Microb Ecol 3(1):1–9. https://doi.org/10.1007/BF02011449
Hentschel U, Schmid M, Wagner M, Fieseler L, Gernert C, Hacker J (2001) Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the Mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. FEMS Microbiol Ecol 35(3):305–312. https://doi.org/10.1111/j.1574-6941.2001.tb00816.x
Sipkema D, Schippers K, Maalcke WJ, Yang Y, Salim S, Blanch HW (2011) Multiple approaches to enhance the cultivability of bacteria associated with the marine sponge Haliclona (gellius) sp. Appl Environ Microbiol 77(6):2130–2140. https://doi.org/10.1128/AEM.01203-10
Jensen S, Fortunato SA, Hoffmann F, Hoem S, Rapp HT, Øvreås L, Torsvik VL (2016) The relative abundance and transcriptional activity of marine sponge-associated microorganisms emphasizing groups involved in sulfur cycle. Microb Ecol 73(3):668–676. https://doi.org/10.1007/s00248-016-0836-3
Lavy A, Keren R, Yu K, Thomas BC, Alvarez-Cohen L, Banfield JF, Ilan M (2018) A novel Chromatiales bacterium is a potential sulfide oxidizer in multiple orders of marine sponges. Environ Microbiol 20(2):800–814. https://doi.org/10.1111/1462-2920
Santos OC, Pontes PV, Santos JF, Muricy G, Giambiagi-deMarval M, Laport MS (2010) Isolation, characterization and phylogeny of sponge-associated bacteria with antimicrobial activities from Brazil. Res Microbiol 161(7):604–612. https://doi.org/10.1016/j.resmic.2010.05.013
Santos-Gandelman JF, Santos OC, Pontes PV, Andrade CL, Korenblum E, Muricy G, Giambiagi-Demarval M, Laport MS (2013) Characterization of cultivable bacteria from Brazilian sponges. Mar Biotechnol (NY) 15(6):668–676. https://doi.org/10.1007/s10126-013-9518-z
Webb MD, Pin C, Peck MW, Stringer SC (2007) Historical and Contemporary NaCl concentrations affect the duration and distribution of lag times from individual spores of nonproteolytic Clostridium botulinum. Appl Environ Microbiol 73(7):2118–2127. https://doi.org/10.1128/AEM.01744-06
Alain K, Querellou J (2009) Cultivating the uncultured: limits, advances and future challenges. Extremophiles 13(4):583–594. https://doi.org/10.1007/s00792-009-0261-3
Nagy E, Boyanova L, US Justesen, ESCMID Study Group of Anaerobic Infections (2018) How to isolate, identify and determine antimicrobial susceptibility of anaerobic bacteria in routine laboratories? Clin Microbiol Infect. https://doi.org/10.1016/j.cmi.2018.02.008
Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P Cai J, Hippe H, Farrow JA (1994) The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol 44(4):812–826. https://doi.org/10.1099/00207713-44-4-812
Yutin N, Galperin MY (2013) A genomic update on clostridial phylogeny: Gram-negative spore-formers and other misplaced clostridia. Environ Microbiol 15(10):2631–2641. https://doi.org/10.1111/1462-2920
Sasi Jyothsna T, Tushar L, Sasikala C, Ramana C (2016) Paraclostridium benzoelyticum gen nov., sp. nov., isolated from marine sediment and reclassification of Clostridium bifermentans as Paraclostridium bifermentans comb nov. Proposal of a new genus Paeniclostridium gen. nov. to accommodate Clostridium sordellii and Clostridium ghonii. Int J Syst Evol Microbiol 66(3):1268–1274. https://doi.org/10.1099/ijsem.0.000874
Lawson PA, Citron DM, Tyrrell KL, Finegold SM (2016) Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) Prévot 1938. Anaerobe 40:95–99. https://doi.org/10.1016/j.anaerobe.2016.06.008
Galperin MY, Bover V, Tolstoy I, Yutin N (2016) Phylogenomic analysis of the family Peptostreptococcaceae (Clostridium cluster XI) and proposal for reclassification of Clostridium litorale (Fendrich et al. 1991) and Eubacterium acidaminophilum (Zindel et al. 1989) as Peptoclostridium litorale gen. nov. comb. nov. and Peptoclostridium acidaminophilum comb. nov. Int J Syst Evol Microbiol 66(12):5506–5513. https://doi.org/10.1099/ijsem.0.001548
Singhal N, Kumar M, Kanaujia PK, Virdi JS (2015) MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol 6:791. https://doi.org/10.3389/fmicb.2015.00791
Schaumann R, Dallacker-Losensky K, Rosenkranz C, Genzel GH, Stîngu CS, Schellenberger W, Schulz-Stübner S, Rodloff AC, Eschrich (2018) Discrimination of human pathogen Clostridium species especially of the heterogeneous C. sporogenes and C. botulinum by MALDI-TOF Mass Spectrometry. Curr Microbiol 75(11):1506–1515. https://doi.org/10.1007/s00284-018-1552-7
Dieckmann R, Graeber I, Kaesler I, Szewzyk U, von Döhren H (2005) Rapid screening and dereplication of bacterial isolates from marine sponges of the Sula Ridge by Intact-Cell-MALDI-TOF mass spectrometry (ICM-MS). Appl Microbiol Biotechnol 67:539–548. https://doi.org/10.1007/s00253-004-1812-2
Dieckmann R, Strauch E, Alter T (2010) Rapid identification and characterization of Vibrio species using whole-cell MALDI-TOF mass spectrometry. J Appl Microbiol 109:199–211. https://doi.org/10.1111/j.1365-2672.2009.04647.x
Emami K, Nelson A, Hack E, Zhang J, Green DH, Caldwell GS, Mesbahi E (2016) MALDI-TOF mass spectrometry discriminates known species and marine environmental isolates of Pseudoalteromonas. Front Microbiol 7:104. https://doi.org/10.3389/fmicb.2016.00104
Ng HJ, Webb HK, Crawford RJ, Malherbe F, Butt H, Knight R, Mikhailov VV, Ivanova EP (2013) Updating the taxonomic toolbox: classification of Alteromonas spp. using multilocus phylogenetic analysis and MALDI-TOF mass spectrometry. Antonie van Leeuwenhoek 103(2):265–275. https://doi.org/10.1007/s10482-012-9807-y
Rahi P, Prakash O, Shouche YS (2016) Matrix-assisted Laser Desorption/Ionization Time-of-Flight Mass-Spectrometry (MALDI-TOF MS) based microbial identifications: challenges and scopes for microbial ecologists. Front Microbiol 7:1359. https://doi.org/10.3389/fmicb.2016.01359
Tushar L, Sasi Jyothsna TS, Sasikala C, Ramana CV (2015) Draft genome sequence of antimicrobial-producing Clostridium sp. JC272, isolated from marine sediment. Genome Announc 3(3):e00650-15. https://doi.org/10.1128/genomea.00650-15
Kok MS (2015) An integrated approach: advances in the use of Clostridium for biofuel. Biotechnol Genet Eng Rev 31(1–2):69–81. https://doi.org/10.1080/02648725.2016.1168075
Leja K, Myszka K, Olejnik-Schmidt AK, Juzwa W, Czaczyk K (2014) Selection and characterization of Clostridium bifermentans strains from natural environment capable of producing 1,3-propanediol under microaerophilic conditions. AJMR 8(11):1187–1197. https://doi.org/10.5897/AJMR213.6516
Myszka K, Leja K, Olejnik-Schmidt AK, Czaczyk K (2012) Isolation process of industrially useful Clostridium bifermentans from natural samples. J Biosci Bioeng 113(5):631–633. https://doi.org/10.1016/j.jbiosc.2012.01.003
Leja K, Myszka K, Czaczyk K (2013) The ability of Clostridium bifermentans strains to lactic acid biosynthesis in various environmental conditions. Springerplus 2(1):44. https://doi.org/10.1186/2193-1801-2-44
Zhang S, Kim TH, Lee Y, Hwang SJ (2012) Effects of VFAs concentration on bio-hydrogen production with Clostridium bifermentans 3AT-ma. Energia Procedia 14:518–523. https://doi.org/10.1016/j.egypro.2011.12.968
Acknowledgments
The research for this present work was financially supported by Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ, Grant Numbers E-26/102.970/2012 and E-26/203.320/2017), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior Programa de Excelência Acadêmica (CAPES ProEx Grant Number 23038.002486/2018-26) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Grant Numbers 448625/2014-8 and 304477/2015-0). MDC was recipient of undergraduate scholarship from CNPq [PIBIC-UFRJ] and BFRO is recipient of doctoral scholarship from CNPq [Grant 140840/2018-4]. GM thanks CAPES, CNPq and FAPERJ for grants and fellowships. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
de Oliveira, B.F.R., Cavalcanti, M.D., de Oliveira Nunes, S. et al. Paraclostridium is the Main Genus of Anaerobic Bacteria Isolated from New Species of the Marine Sponge Plakina in the Brazilian Southeast Coast. Curr Microbiol 76, 713–722 (2019). https://doi.org/10.1007/s00284-019-01684-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-019-01684-x